Abstract
Several lines of evidence have suggested that the long arm of chromosome 12 may carry a tumor-suppressor gene(s) that plays a role in pancreatic ductal carcinogenesis. We have previously found a significant association between loss of heterozygosity of the 12q arm and a poor prognosis in pancreatic cancer patients. In this study, we introduced a normal copy of chromosome 12 into some pancreatic ductal carcinoma cells. Both anchorage-dependent and -independent proliferations as well as invasiveness were similar throughout the hybrid clones when compared with their corresponding parental cells. In sharp contrast, significant suppression of tumorigenesis was observed after inoculation of the hybrid clones into nude mice. Measurements made up to 1 month later showed that there was a significant delay in the growth of tumors into which the introduced normal copy of chromosome 12 had been restored. More significantly, using our dorsal skin chamber and an intravital microscopy system experiment in SCID mice, we demonstrated and visualized directly that implantation of the hybrids failed to promote the angiogenic phenotype encountered in the parental cells. Gene expression profiling using the complementary DNA microarray system identified a set of 24 genes differentially expressed between the hybrids and parental cells. An additional set of 18 genes was also identified that were differentially expressed between the hybrid clone that lost its growth-suppression activity and one that retained such activity. Another set of 25 genes mapped on 12q was detected that showed high expression levels in the hybrid clones retaining growth-suppressive activity. In summary, this study provides the first functional evidence of the existence of an additional tumor-suppressor gene(s) on chromosome 12, whose absence is responsible for the pathogenesis in pancreatic ductal carcinogenesis.
Similar content being viewed by others
Main
Ductal adenocarcinoma is the most frequent malignancy arising in the pancreas. Although the incidence of this disease is only 3.6% of the cancer cases in Japan, the number of cancer deaths caused by this disease accounts for up to 6.4% of the total (http://www.ncc.go.jp/en/statistics/2001/index.html). The mean 5-year survival rate of this disease is poor; it is below 5% in Japan (http://www.mc.pref.osaka.jp/ocr_e/ocr/index.html#survival) and worldwide.1 This poor prognosis is partly due to the lack of symptoms arising only at the late stage; nearly 80% of pancreatic cancer patients already harbor metastases at the time of diagnosis. Detection of small, resectable cancers would improve the outcome of this deadly disease;2 but the optimal approach to early detection of pancreatic cancer has not yet been established. Thus, acquisitions of efficient approaches for accurate detection at the earliest stages as well as development of efficient methods for treatment are among the tasks with the highest priority in conquering pancreatic cancer.
Tumor-suppressor genes (TSGs) and their products are attractive candidates as molecular targets for early genetic diagnosis because their functional losses should be followed by switching toward a malignant phenotype. Moreover, there is the possibility of inventing valuable techniques for clinical management of this disease by supplementation of the lost functions of TSGs. Despite the continuous progress in molecular biology, the genetic events involved in the initiation and progression of pancreatic ductal adenocarcinoma remain largely unclear. Cytogenetic, allelotype, and somatic cell hybrid studies in human cancers have suggested that chromosome 12 may carry a TSG(s) that plays a role in the carcinogenises of prostate,3, 4 stomach,5, 6, 7 male germ cells,8 and pancreas.9, 10, 11 Furthermore, we previously demonstrated that 12q-loss of heterozygosity (LOH) is significantly associated with a poor prognosis in patients with pancreatic cancer.12 DUSP6 on 12q was found to be inactivated in pancreatic cancer,13 and introduction of this gene-induced apoptosis.14 However, no structural abnormality was observed in this gene, and its localization was outside the smallest region of overlap (SRO). Hence, there is a possibility of the localization of an unknown TSG(s) on 12q that is associated with a poor prognosis in pancreatic cancer patients. To address this possibility and to isolate and characterize the TSG(s) on 12q, we first tried to demonstrate the factor on chromosome 12 that harbored tumor-suppressor activity by means of introduction of a normal copy of chromosome 12.
Materials and methods
Cell Lines
The pancreatic cancer cell lines used were PCI-35 and MIAPaCa2; the former is a generous gift from Dr Hiroshi Ishikura at Hokkaido University, and the latter was purchased from the American Type Culture Collection (Manassas, VA, USA). Chromosome 12 was introduced in these cells by the microcell-mediated chromosome transfer (MMCT) method (see below). Cells were cultured according to the protocols of the suppliers. The parental cell lines were previously well mutationally characterized.15 The normal human fibroblast cell line MRC-5 (American Type Culture Collection) and the mouse A9 cell line (provided by Japanese Cancer Research Resources Bank) were maintained according to the suppliers' protocols. All cells were routinely monitored for Mycoplasma as well as for mouse hepatitis, Sendai, and pneumonia viruses and were consistently negative.
MMCT
MRC-5 fibroblast cells were transfected with pSV2neo plasmid DNA and then selected in DMEM medium containing 400 μg/ml G418 (GibcoBRL, Grand Island, NY, USA). Cell hybrids of G418-resistant human fibroblast cells and mouse A9 cells were fused, selected, and pooled as described elsewhere.16 MMCT experiments were performed as described16, 17 using as donors A9H(12) hybrids containing an MRC5 human chromosome 12 tagged with a neomycin-resistance gene, thus allowing clonal selection and expansion in medium containing 400 μg/ml of G418. The resulting final hybrids, five stable clones for each recipient, were named as follows: PCI-35H(12)-1 and -2, and MIAPaCa2H(12)-1 through -3, respectively.
Microsatellite Analysis
Genomic DNA from the A9H(12), parental cell lines, their hybrids, and the corresponding nude mice tumors was analyzed with highly polymorphic microsatellite markers, as described previously.9 A panel of microsatellite markers was selected that spaced at approximately 10-cM intervals along the long arm of chromosome 12 as follows: D12S1701 (12q12), D12S88 (12q21), D12S1719 (12q21), D12S360 (12q22), D12S78 (12q23), and D12S366 (12q24). D12S336 on 12p was also used as the control for the short arm marker. Nucleotide sequences of the markers and conditions for PCR have been described previously.18 The PCR products were separated by running in 6% polyacrylamide/8 M urea/32% formamide gel, followed by fixation in 5% acetic acid/5% methanol for 30 min, drying on a 3 mm filter paper (Whatman Inc., Clifton, NJ, USA) and autoradiography. For each marker, two independent PCR amplifications labelling forward and reverse primers, respectively, were carried out to confirm the results.
Fluorescence In Situ Hybridization (FISH)
FISH analysis was carried out as previously described.10 Briefly, the parental cells and their hybrids were prepared in a metaphase spread by hypotonic treatment and fixation in Carnoy's solution. Dual-color FISH was performed by using two different regional probes for 12q21: b605B21 and b759H8.19 As the corresponding centromere-specific probe, we used α12H8 corresponding to D12Z3 (purchased from ATCC, Rockville, MD, USA). BAC DNAs were labelled with biotin–16-dUTP (green signals), and the centromeric probe was labelled with digoxigenin–11-dUTP (red signals). Fluorescence detection of the signals was performed with antidigoxigenin–tetramethylrhodamine isothiocyanate (TRITC) and avidin–fluorescein isothiocyanate (FITC) (Boehringer Mannheim, Mannheim, Germany) followed by counterstaining using 4′,6-diamino-2 phenylindole (DAPI) in an antifade solution. At least 100 nuclei for every spread were analyzed, and an average TRITC/FITC ratio profile was estimated.
Proliferation Assays
Anchorage-dependent proliferation was monitored by an MTT assay for 5 days in the absence of G418, and a daily proliferation index (PI) was calculated for each parental and corresponding hybrid cell line by the methods described by van Golen et al.20 In all assays, 1000 cells in 100 μl suspension of each cell type were plated and incubated in wells of five flat-bottomed 96-well plates. The conversion of MTT to formazan dye was spectrometrically measured for absorbance at 590 nm using a multiwell plate ImmunoReader System (Molecular Dynamics, Inc., Sunnyvale, CA, USA). All experiments were performed in duplicates of eight and repeated at least twice. For each cell line, the PI was estimated as previously described.21 Data from two independent experiments were pooled, averaged, and then statistically analyzed.
For anchorage-independent proliferation, 10 000 cells of each parental and hybrid line were plated in 1 ml medium containing 0.3% Bacto-agar (Becton Dickinson, Sparks, MD, USA) with 10% fetal bovine serum (FBS) as an upper layer into 30-mm dishes. Another 1 ml medium with 0.7% Bacto-agar and 10% FBS was used for the bottom layer. Dishes were maintained in a humidified 5% CO2 atmosphere at 37°C and fed biweekly with 0.3 ml medium. After 21 days, 0.3 ml of 1 mg/ml INT (2-[4-iodophenyl]-3-[4-nitrophenyl]-5-phenyl-2H-tetrazolium chloride) solution (Dojindo Laboratories, Kumamoto, Japan) was added in each dish and further incubated for another 3 h. The viable, red-stained colonies were photographed using a Zeiss microscope (Carl Zeiss, Göttingen, Germany). Both colony number and size were measured and averaged on three randomly chosen photographs from each plate by using public domain NIH1.62 software. Every anchorage-independent growth was assessed in triplicate by two independent experiments.
Tumorigenicity in SCID Mice
Female SCID mice, 5 weeks old, purchased from Clea Japan Inc. (Tokyo, Japan) were maintained under pathogen-free conditions and used in accordance with NIH and Tohoku University Medical School institutional guidelines. Logarithmically growing cells trypsinized from subconfluent monolayers were suspended in medium containing 25% Matrigel Growth Factor Reduced (Becton Dickinson Labware, Franklin Lakes, NJ, USA) at a density of 1x107 cells/ml. For each inoculation, 3 × 106 cells in 0.3 ml suspension were injected s.c. into the hind flanks of nude mice. For every pair of cells, inoculations were performed in three mice. The tumor volume was estimated by the formula: V=0.4Dd2 (V=tumor volume, D=longitudinal diameter, and d=latitudinal diameter) at the time of biweekly measurements. Data from two independent experiments were pooled for statistical analysis.
In Vivo Microscopy
The dorsal transparent chamber and in vivo microscopy system are described elsewhere.22 A total of 1 × 106 cells of either parental cells or their corresponding hybrids were implanted into the mice. Tumor vessel formation was observed for 3 weeks after tumor cell implantation. Images were captured by a CCD camera (TEC-470 Optronics Co., Chelmsford, MA, USA) attached to a microscope (Nikon, Tokyo, Japan), and recorded on a Super VHS video recorder (Victor, Kanagawa, Japan). Finally, the images were analyzed and prepared off-line using Avid VideoShop 3.0.2 (Avid Technology Inc., Tewksbury, MA, USA) and Adobe Photoshop 5.0.2 (Adobe Systems Inc., San Jose, CA, USA) software.
Immunohistochemistry
Resected specimens were fixed with 4% paraformaldehyde solution overnight. After embedding in OCT compound, the specimens were frozen at −80°C. We used 4 μm sections from frozen specimens for immunohistochemical staining. Anti-mouse CD31 antibody (BD Biosciences-Pharmingen, San Diego, CA, USA) diluted 100-fold in PBS was used as the primary antibody and incubated for 1 h at room temperature. Then the peroxidase-conjugated anti-rat IgG antibody (BD Biosciencies-Pharmingen) was used for the secondary antibody reaction and incubated for 30 min at room temperature, followed by a reaction with AEC reagents (Vector Laboratories, Burlingame, CA, USA) for 10 min at room temperature. AEC reagents were used as chromogens, and hematoxylin was used for counterstaining.
Microarray Analysis
Total RNAs were extracted from the cultured hybrids and their corresponding parental cells using an RNeasy Midi Kit (QIAGEN, Valencia, CA, USA), and the messenger RNAs (mRNAs) were refined from the total RNAs with an Oligotex-dt30 mRNA purification kit (TAKARA, Kyoto, Japan) according to the suppliers' protocols. Cy3- and Cy5-labelled complementary DNA (cDNA) probes for hybridization were prepared from refined mRNAs with a CyScribe First-Strand cDNA Labelling kit and a CyScribe GFX Purification kit (Amersham Biosciences, Piscataway, NJ, USA) according to the supplier's protocols. We used cDNA microarray slides fabricated by spotting 23 040 unique cDNAs purchased from Amersham Biosciences on Type 7 slides using the Gen III Array Spotter (Amersham Biosciences). The cDNAs were selected from UniGene database,23 including expressed sequence tags (ESTs), and prepared by PCR-amplification with unique primers. The Lucidea Microarray ScoreCard system (Amersham Biosciences) containing 32 control samples including 11 human housekeeping genes and several artificial cDNAs was used for checking dynamic range and variations of signal intensities according to the supplier's instructions. Each cDNA was spotted in duplicate. Each 10 pmol of labelled probes were hybridized by using Automated Slide Processor according to the supplier's instructions (Amersham Biosciences). The hybridized slides were scanned with a GenePix 4000A scanner (Axon Instruments, Union City, CA, USA). The scanned image was converted to intensity values using GenePix Pro software (Axon Instruments). Duplicated hybridization experiments were carried out for all samples to confirm the results. Standardization of signals was carried out by using the Lucida Microarray ScoreCard software according to the supplier's instructions. The standardized data were analyzed using the GeneSpring software (Silicone Genetics, Red Wood City, CA, USA). Interspot normalization was carried out by dividing sample signal values by control channel values. Interslide normalization was carried out by dividing each signal value by median values of 11 housekeeping genes. We selected genes whose expression showed more than 1.5-fold difference, either higher or lower, with a statistical significance of less than 0.05 in probabilities by ANOVA and t-test. Significant differences between parental cells and hybrids were accepted to be less than 0.05 by ANOVA and t-test as provided in the software. Annotated information about genes was obtained by using the GeneSpider program provided in the software. Grouping of genes according to ontology was carried out by using the GeneOntology program provided in the software.
Reverse Transcription-PCR
Total RNAs extracted from cell pellets were used for reverse transcription reactions with SuperScript II RNase H-reverse transcriptase (Invitrogen, Inc., San Diego, CA, USA) according to the method described previously.24 For semiquantitative reverse transcription-PCR, concentrations of template cDNAs were adjusted to give the same quantity by β2-microglobulin mRNA measured by ethidium bromide staining in agarose gel electrophoresis. Sequences of primer and optimized conditions for reactions are available upon request. For quantitative RT-PCR, we designed specific primers and fluorescence-labelled probes for the RAB21 with the primer express software (Applied Biosystems, Foster City, CA, USA). Each cDNA was subjected to 40 cycles of the PCR using the ABI PRISM 7700 (Applied Biosystems) according to the supplier's instructions.
Statistical Analysis
All experiments were performed in duplicate or triplicate. A two-tailed Student's t-test was performed by using the StatView software 5.0 (SAS Institute Inc., Cary, NC, USA) to determine the statistical significance of differences. The level of significance was established at P<0.05.
Results
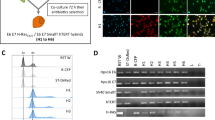
In this study, we utilized the technique of MMCT to introduce a normal copy of human chromosome 12 individually into two pancreatic cancer cell lines, MIAPaCa2 and PCI-35. In a previous investigation, MIAPaCa2 showed a loss of chromosome 12q arm, whereas PCI-35 did not.18 We established three independent hybrid clones for MIAPaCa2, MIAPaCa2H(12)-1, -2 and -3, and two independent clones for PCI-35, PCI-35H(12)-1 and -2. To elucidate portions of retained alleles, we performed a microsatellite analysis using a panel of highly polymorphic markers on chromosome 12. However, we could not distinguish between existing alleles and the introduced alleles for most of markers we analyzed because of the few heterozygosities of the microsatellite markers (data not shown). Therefore, we performed a dual-color FISH analysis in duplicate to monitor both the number of introduced chromosomes and the percentage of cells maintaining the introduced chromosome. Markers used were as below: chromosome 12 centromere, 12q21, and 12q23.1. These markers were selected because their high frequencies of losses in primary pancreatic cancer have been reported.18 Previous works reported that MIAPaCa2 was hypotriploid with losses of 12q21 and 12q23.1, whereas PCI-35 was mainly hypotetraploid without those losses.18, 25 Consistent with the previous results, our FISH analyses, as shown in Figure 1, indicated losses of 12q21 (b759H8) and 12q23.1 (b339F2) in the majority of cells of MIAPaCa2 (Table 1). On the other hand, we found additional signals in the hybrid cells indicating one more copy of portions of the centromere, 12q21 and 12q23.1, as shown in Figure 1; the most frequently observed signal pattern for cen/12q21 or cen/12q23.1 was 3/2 in MIAPaCa2, whereas that in the hybrid was 4/3. Notably, we detected loss of the introduced chromosome in the MIAPaCa2H(12)-2, showing the 3/2 pattern in the majority of cells (P<0.05), although the cells maintained resistance to G418. Confirming our previous results,18 PCI-35 shows an apparently nondefective status of 12q: predominant ratios were 4/4 and 5/5 in parental and PCI-35H(12) cells, respectively. These results are summarized in Table 1. As clearly indicated in Table 1, the hybrid cells we employed were mixed populations of various status of the transferred chromosome because the transferred chromosomes are not stable and sometimes partial loss occurs during the course of serial passages. Each population may express different phenotypes, but we could only observe the mixed phenotypes because of technical difficulties in isolating a pure population.
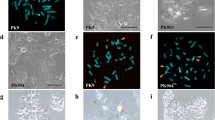
Representative images of FISH analysis for pancreatic cancer cells and their chromosome 12-hybrid clones. Red signals, fluorescence detection with TRITC for the centromere of chromosome 12 (pα12H8); green signals, fluorescence detection with FITC for D12S88 localizing at 12q21 (b759H8). (a) Chromosome spread of the metaphase of normal cells. (b–e) Interphase nuclei of MIAPaCa2 (b), MIAPaCa2H(12)-3 (c), PCI-35 (d), and PCI-35H(12)-1 (e).
Once the presence of the introduced chromosome 12 in the hybrids was verified, we estimated their in vitro proliferation in either an anchorage-dependent or -independent manner. The results as outlined in Figure 2 show that the in vitro growth of the hybrid clones was not significantly different from that of parental cells.
In vitro anchorage-dependent proliferation assay. The data were plotted in logarithmic scale. (a) PCI-35 (PW) and PCI-35 hybrid clones (P1 and P2). (b) MIAPaCa2 (MW) and MIAPaCa2 hybrid clones (M1, M2 and M3). (c) In vitro anchorage-independent proliferation assay. Each solid bar indicates the number of the colonies in the medium with Bacto-agar after 1 month of culture of cells of PCI-35 (PW), PCI-35 hybrid clones (P1 and P2), MIAPaCa2 (MW), and MIAPaCa2 hybrid clones (M1, M2 and M3).
Next, we examined the in vivo tumorigenesis phenotypes of the hybrid cells by inoculating them into SCID mice and comparing them with parental cells. In order to shorten tumor latency and enhance tumor growth, we mixed the cells in a suspension containing Matrigel extract. As shown in Figure 3, hybrids MIAPaCa2H(12)-1 and -3 showed significant reductions in tumor volume and a longer latency when compared with their parental cells. MIAPaCa2H(12)-2 and the hybrid clones derived from PCI-35 did not show any significant reduction in tumor volume. Microscopically, tumors generated of the hybrids MIAPaCa2H(12)-1 and -3 tended to form fibrotic changes and showed significant reductions in vessel number as proven by the quantitative vessel counting assay comparing numbers with those in tumors grown from the parental cells (Figure 4). In addition, to compare angiogenesis between MIAPaCa2 and the hybrids, we monitored tumor vessel formation for 3 weeks after tumor cell implantation in a dorsal transparent chamber and an in vivo microscopy system. An intense blood vessel formation implying active vascularization was observed in tumors derived from MIAPaCa2 parental cells. On the other hand, tumors of hybrid clones showed very sparse vessel formation (see Figure 5). These results indicated that some of the hybrid cells had a significant reduction in in vivo tumorigenic activity, which could be accounted for by the suppression of angiogenesis.
In vivo tumorigenic assay employing a total of 3 × 106 tumor cells inoculated into subcutaneum of SCID mice. (a, b) Representative pictures taken at 1 month after the inoculation: (a) PCI-35 (PW1) and PCI-35H(12)-1 (P1), (b) MIAPaCa2 (MW3) and MIAPaCa2H(12)-3 (M3). (c, d) Growth curves of the inoculated tumors. The volume of tumors were plotted in logarithmic scale. (c) PCI-35 clones (PW1 and PW2) and PCI-35 hybrid clones (P1 and P2); (d) MIAPaCa2 clones (MW1, MW2 and MW3) and MIAPaCa2 hybrid clones (M1, M2 and M3). Asterisks denote statistically significant differences (P<0.05).
(a–d) Immunohistochemistry employing with anti-CD31 antibody for specimens of inoculated tumors of MIAPaCa2 (a), MIAPaCa2H(12)-3 (b), PCI-35 (c) and PCI-35H(12)-1 (d) (× 400). (e, f) Quantification of CD31-positive areas by counting pixels in digitally imported images. (e) MIAPaCa2 (MW) and MIAPaCa2 hybrid clones (M1, M2 and M3). (f) PCI-35 (PW) and PCI-35 hybrid clones (P1 and P2). Asterisks denote statistically significant differences (P<0.05).
Next, we performed a cDNA microarray analysis to determine the differences in gene expression profiles between the parental MIAPaCa2 and its hybrid clones, because significant phenotypic differences were observed in this set. We employed a cDNA microarray platform consisting of 23 040 genes commercially available from Amersham Biosciences (Piscataway, NJ, USA) and performed a comparative hybridization analysis between the parental cells and their hybrids with suppressed tumorigenicity phenotype. The results give us information about differentially expressed genes, theoretically caused by the introduction of chromosome 12. We selected genes with differential values of more than 1.5-fold and showed statistically significant differences. Among the results, we found that 24 genes met the criteria (see Table 2). Predicted functions were annotated based on the Gene Ontology database.
Next, we compared the expression profiles between MIAPaCa2H(12)-2 and MIAPaCa2H(12)-3. Although both were MMCT hybrid clones, the former lost its growth-suppressive activity, while the latter retained it after inoculation into SCID mice. This comparison may give significant information of genes accounting for the tumor-suppressive phenotype without a noise of MMCT technique itself. We found that 18 genes showed more than a 1.5-fold difference in expression level, which is a statistically significant difference (Table 3). These genes could account for the differences in tumorigenic and angiogenic phenotypes between the clones.
Not only genes on chromosome 12 that were expressed differentially beyond our criteria but also those that were expressed below the criteria could produce the tumor-suppressive phenotype, because the addition of one allele to the existing three alleles on chromosome 12 may not result in a significant difference in expression levels in some genes, especially in those supposed to be functionally altered by structural alteration. Therefore, we searched for all expressed genes beyond background levels in hybrids of MIAPaCa2H(12)-1 and -3, both of which showed the suppressed tumorigenic phenotype, and found 25 genes on chromosome 12 according to the annotated information as listed in Table 4.
We validated the results of alterations of expressions detected in the microarray experiment by the semiquantitative RT-PCR method (see Figure 6). Although the results of semiquantitative RT-PCR were not completely consistent with the corresponding data of microarray experiment in the magnitude of change in expression level, the direction of change, either upregulation or downregulation, in each case was retained. Among these, the RAB21 gene, one of the candidate genes selected by microarray analysis and located on chromosome arm 12q, showed a higher expression in MIAPaCa2 hybrids than in their parental cells. The results of microarray analysis were reconfirmed by the quantitative real-time RT-PCR method (see Figure 7).
Semireal-time quantitative RT-PCR for MIAPaCa2 parental cell and its derived hybrid clones. β2-Microglobulin mRNA was used as the control to adjust the concentrations of template cDNAs. PCR was performed in minimum cycles and results were confirmed by ethidium bromide staining after agarose gel electrophoresis. Lanes MW, MIAPaCa2 parental cell; M1, hybrid clone 1; M2, hybrid clone 2; M3, hybrid clone 3.
Discussion
Several lines of evidence, as we described in the Introduction, have suggested that chromosome 12q may carry a TSG(s) that plays a role in the development and/or progression of pancreatic cancer. We aim to gather functional evidence for the existence of TSG(s) and refine candidate(s) yet to be identified on the 12q arm. We transferred a normal copy of chromosome 12 into pancreatic cancer cell lines by the microcell-mediated chromosome transfer (MMCT) technique16, 17 and analyzed its phenotype. MMCT has been proven to be a useful tool providing functional evidence for identification of TSG in a variety of cancers such as pancreatic cancer,21 colon cancer,26, 27 prostate cancer,28 Wilms' tumor,29 and melanoma.30 This technique also led the way to the isolation of the NBS gene.31
The derived hybrids of chromosome 12 showed clear differences from parental cells not in in vitro but in vivo tumorigenic study. The in vitro studies of anchorage-dependent and -independent cell proliferations showed no remarkable differences. However, the inoculation of the hybrid cells MIAPaCa2H(12) into SCID mice strikingly showed a significant suppression of tumorigenesis when compared with parental cells of MIAPaCa2. The hybrid cells of the PCI-35 lineage did not show such a phenotype. These results were of particular interest because MIAPaCa2 was partially defective for chromosome 12q, but PCI-35 was not.18 These results suggested that newly introduced genes on chromosome 12 overcame defective functions of existing genes in MIAPaCa2 but not in PCI-35. One clone of hybrid of MIAPaCa2, MIAPaCa2H(12)-2, did not show a suppressive phenotype. We suspected that this clone lacked some important portions of the introduced allele. However, we could not detect differences in genotypes regarding chromosome 12 among hybrids derived from MIAPaCa2 in our panel of microsatellite analysis, mainly because of similarities in the number of repeats, which consisted of the microsatellites between the existing alleles and the introduced allele.
We found a remarkable suppression of angiogenesis in and surrounding the inoculated tumors of hybrids in examinations employing quantitative vessel counting with immunohistochemical labelling and an in vivo microscopy system. The suppression of angiogenesis could account for the suppressive phenotype of in vivo tumorigenesis. These facts indicated a potential interposition of putative TSGs at chromosome 12 playing suppressive roles, not in proliferation of the tumor itself in an early phase, but in angiogenesis in a later phase of tumorigenesis. Angiogenesis is a key factor for tumorigenesis, and its suppression plays a major role in a tumor-suppressive activity.32, 33, 34 The suppressive activity of introduction of chromosome 12 in the later phase of pancreatic tumorigenesis could explain our previous finding of significant association of loss of chromosome 12q with poor prognoses in patients with pancreatic cancer.12
We further analyzed a total of 23 040 unique human genes in this study to search for genes closely associated with tumorigenesis by altering expression and successfully grasped gene expression profiles of hybrids in comparison to parental cells. Although the possibility of missing important genes cannot be excluded because of the limited number we examined, about two-thirds of the total human genes that were analyzed, this method is one of the best ways to explore the genes that play important roles in pancreatic carcinogenesis. Using this method, detection of genes that lose their function by structural alterations cannot be detected either. However, this technique enabled us to obtain valuable information from various aspects such as examinations of the TGF-beta/SMAD4 pathway35 or introduction of a DNA methylation inhibitor or a selective COX-2 inhibitor in pancreatic cancer cells.36, 37 Discovery of various overexpressed genes in pancreatic cancer cells was also reported by this method; sea urchin fascin homolog, heat shock protein,38 ABL2, Notch4, SOD1,39 c-myc and Rad51.40 We report herein the first results of microarray analysis of the comparison between parental cells and their hybrids after introduction of chromosome 12 using the MMCT technique.
For analyzing the data of the microarray, we first selected genes whose expressions were significantly different statistically and more than 1.5-fold differentially expressed genes between the parental cell and hybrid clones were picked up. These data gave information about alteration of gene expression by introduction of the additional copy of chromosome 12. According to our FISH results, the majority of hybrid cells harbored one additional copy of the chromosome 12 to tri- and tetraploid cells, MIAPaCa2 and PCI-35, respectively. Although we could not precisely estimate alterations of expressions of genes between parental cells with three or four alleles and cells with one additional allele in them, it is probable that the differences may be small. Therefore, we used only statistical methods for data analysis without cutting off the data in large-fold increases of expression levels as described in other reports; we applied 1.5-fold as the cutoff. We obtained 24 genes that were differentially expressed between the MIAPaCa2 and its derived hybrid clones with suppressed tumorigenicity. These 24 genes are predicted to function in a variety of pathways and situations, potentially indicating complicated molecular networks underlying cellular phenotypes triggered by genes on the transferred chromosome and/or the effect of introduction of one additional allele itself. Since we applied the strict statistical method to select genes to avoid false positives, and because we excluded ESTs without annotated information, the total number of genes listed is not large. Among those selected genes, several interesting genes have been reported in association with cell proliferation. PPP2CA comprises a diverse family of phosphoserine- and phosphothreonine-specific enzymes ubiquitously expressed in eukaryotic cells, and regulates a diverse set of cellular processes such as metabolism, cell cycle, signal transduction, differentiation, and oncogenic transformation.41 CNAP1 is one of the essential components of the chromosome condensation complex in the mitotic process, and a mutant CNAP1 was unable to associate with mitotic chromosomes.42
Gene expression profiles between the two typical hybrid clones of MIAPaCa2, one of which lost growth-suppressive activity (MIAPaCa2H(12)-2) and the other that retained it (MIAPaCa2H(12)-3), could give us valuable information about genes accounting for the difference of phenotype. The 18 selected, differentially expressed genes were scattered on various chromosomes and had a variety of functions; some important clues may be hidden in the function of these genes.
The 25 expressed genes on chromosome 12 in MIAPaCa2H(12)-1 and -3 potentially include genes functioning in the tumor-suppressive pathway in pancreatic cancer. The ranges of expression levels were between 0.31 and 3.86 when compared with parental cells; downregulation of genes on chromosome 12 could be a result of direct or indirect trans-suppression by introduced genes. Some of these 25 genes on chromosome 12 already showed evidence of suppressor activity in pancreatic cancer. TGF pathway components may use a motor protein light chain as a receptor for the recruitment and transport of specific cargo along microtubles.43 PRPN6 encodes the protein tyrosine phosphatase of nonreceptor type 6, which is shown to be suppressed in leukemic cells and correlated with patients' prognosis.44 Among these genes on chromosome 12, RAB21 was upregulated 1.68-fold higher in hybrids, and this fold change was reconfirmed by quantitative PCR. RAB21 is a member of a subfamily of small GTP-binding protein of the Ras superfamily that has been revealed to play a role in the regulation of vesicular transport in polarized intestinal epithelial cells.45 Phenotypes related to the carcinogenesis of this gene are yet to be investigated.
In previous works, we have found that expression of DUSP6 at 12q21–q22 is suppressed in pancreatic cancer cells, and exogenous restoration of the gene revealed a tumor-suppressive phenotype.13, 14 It is notable that suppressed expression of DUSP6 was not recovered after restoration of chromosome 12. Probably, an epigenetic mechanism silenced the DUSP6 gene expression of the newly introduced chromosome 12. The roles of other candidate genes in tumor suppressions remain to be explored.
The hybrid cells revealed the suppressive phenotype of angiogenesis: therefore, molecules predicted to be involved in angiogenic process are of particular interest. The microarray we employed contained several genes related to angiogenesis, including ANGPT2, TNFSF12, SH2D2A, ANG, ANPEP, VEGFC, and PGF. We found that these genes were expressed at levels less than the background in both parental and hybrid cells. Therefore, there is little possibility, if any, that they play a role in the angiogenic phenotypes observed in the present study. Further detailed examination of other angiogenesis-related molecules is needed.
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Parkin DM, Pisani P, Ferlay J . Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer 1999;80:827–841.
Ariyama J, Suyama M, Ogawa K, et al. The detection and prognosis of small pancreatic carcinoma. Int J Pancreatol 1990;7:37–47.
Berube NG, Speevak MD, Chevrette M . Suppression of tumorigenicity of human prostate cancer cells by introduction of human chromosome del(12)(q13). Cancer Res 1994;54:3077–3081.
Luu HH, Zagaja GP, Dubauskas Z, et al. Identification of a novel metastasis-suppressor region on human chromosome 12. Cancer Res 1998;58:3561–3565.
Fey MF, Hesketh C, Wainscoat JS, et al. Clonal allele loss in gastrointestinal cancers. Br J Cancer 1989;59:750–754.
Sano T, Tsujino T, Yoshida K, et al. Frequent loss of heterozygosity on chromosomes 1q, 5q, and 17p in human gastric carcinomas. Cancer Res 1991;51:2926–2931.
Schneider BG, Pulitzer DR, Brown RD, et al. Allelic imbalance in gastric cancer: an affected site on chromosome arm 3p. Genes Chromosomes Cancer 1995;13:263–271.
Murty VV, Houldsworth J, Baldwin S, et al. Allelic deletions in the long arm of chromosome 12 identify sites of candidate tumor suppressor genes in male germ cell tumors. Proc Natl Acad Sci USA 1992;89:11006–11010.
Kimura M, Abe T, Sunamura M, et al. Detailed deletion mapping on chromosome arm 12q in human pancreatic adenocarcinoma: identification of a 1-cM region of common allelic loss. Genes Chromosomes Cancer 1996;17:88–93.
Fukushige S, Waldman FM, Kimura M, et al. Frequent gain of copy number on the long arm of chromosome 20 in human pancreatic adenocarcinoma. Genes Chromosomes Cancer 1997;19:161–169.
Heidenblad M, Jonson T, Mahlamaki EH, et al. Detailed genomic mapping and expression analyses of 12p amplifications in pancreatic carcinomas reveal a 3.5-Mb target region for amplification. Genes Chromosomes Cancer 2002;34:211–223.
Yatsuoka T, Sunamura M, Furukawa T, et al. Association of poor prognosis with loss of 12q, 17p, and 18q, and concordant loss of 6q/17p and 12q/18q in human pancreatic ductal adenocarcinoma. Am J Gastroenterol 2000;95:2080–2085.
Furukawa T, Yatsuoka T, Youssef EM, et al. Genomic analysis of DUSP6, a dual specificity MAP kinase phosphatase, in pancreatic cancer. Cytogenet Cell Genet 1998;82:156–159.
Furukawa T, Sunamura M, Motoi F, et al. Potential tumor suppressive pathway involving DUSP6/MKP-3 in pancreatic cancer. Am J Pathol 2003;162:1807–1815.
Sun C, Yamato T, Furukawa T, et al. Characterization of the mutations of the K-ras, p53, p16, and SMAD4 genes in 15 human pancreatic cancer cell lines. Oncol Rep 2001;8:89–92.
Koi M, Morita H, Yamada H, et al. Normal human chromosome 11 suppresses tumorigenicity of human cervical tumor cell line SiHa. Mol Carcinog 1989;2:12–21.
Fournier RE, Ruddle FH . Stable association of the human transgenome and host murine chromosomes demonstrated with trispecific microcell hybrids. Proc Natl Acad Sci USA 1977;74:3937–3941.
Kimura M, Furukawa T, Abe T, et al. Identification of two common regions of allelic loss in chromosome arm 12q in human pancreatic cancer. Cancer Res 1998;58:2456–2460.
Youssef EM, Kaneko K, Yatsuoka T, et al. Human BAC contig covering the deleted region in pancreatic cancer at 12q21. DNA Seq 2001;11:541–546.
van Golen KL, Wu ZF, Qiao XT, et al. RhoC GTPase, a novel transforming oncogene for human mammary epithelial cells that partially recapitulates the inflammatory breast cancer phenotype. Cancer Res 2000;60:5832–5838.
Lefter LP, Furukawa T, Sunamura M, et al. Suppression of the tumorigenic phenotype by chromosome 18 transfer into pancreatic cancer cell lines. Genes Chromosomes Cancer 2002;34:234–242.
Duda DG, Sunamura M, Lozonschi L, et al. Direct in vitro evidence and in vivo analysis of the antiangiogenesis effects of interleukin 12. Cancer Res 2000;60:1111–1116.
Schuler GD, Boguski MS, Stewart EA, et al. A gene map of the human genome. Science 1996;274:540–546.
Mori Y, Shiwaku H, Fukushige S, et al. Alternative splicing of hMSH2 in normal human tissues. Hum Genet 1997;99:590–595.
Yunis AA, Arimura GK, Russin DJ . Human pancreatic carcinoma (MIA PaCa-2) in continuous culture: sensitivity to asparaginase. Int J Cancer 1977;19:218–235.
Tanaka K, Oshimura M, Kikuchi R, et al. Suppression of tumorigenicity in human colon carcinoma cells by introduction of normal chromosome 5 or 18. Nature 1991;349:340–342.
Tanaka K, Yanoshita R, Konishi M, et al. Suppression of tumorigenicity in human colon carcinoma cells by introduction of normal chromosome 1p36 region. Oncogene 1993;8:2253–2258.
Padalecki SS, Johnson-Pais TL, Killary AM, et al. Chromosome 18 suppresses the tumorigenicity of prostate cancer cells. Genes Chromosomes Cancer 2001;30:221–229.
Weissman BE, Saxon PJ, Pasquale SR, et al. Introduction of a normal human chromosome 11 into a Wilms' tumor cell line controls its tumorigenic expression. Science 1987;236:175–180.
Trent JM, Stanbridge EJ, McBride HL, et al. Tumorigenicity in human melanoma cell lines controlled by introduction of human chromosome 6. Science 1990;247:568–571.
Matsuura S, Tauchi H, Nakamura A, et al. Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet 1998;19:179–181.
Folkman J . Tumor angiogenesis: therapeutic implications. N Engl J Med 1971;285:1182–1186.
St Croix B, Rago C, Velculescu V, et al. Genes expressed in human tumor endothelium. Science 2000;289:1197–1202.
Bergers G, Javaherian K, Lo KM, et al. Effects of angiogenesis inhibitors on multistage carcinogenesis in mice. Science 1999;284:808–812.
Holloway S, Davis M, Jaber R, et al. A clinically relevant model of human pancreatic adenocarcinoma identifies patterns of metastasis associated with alterations of the TGF-beta/Smad4 signaling pathway. Int J Gastrointest Cancer 2003;33:61–69.
Tseng WW, Deganutti A, Chen MN, et al. Selective cyclooxygenase-2 inhibitor rofecoxib (Vioxx) induces expression of cell cycle arrest genes and slows tumor growth in human pancreatic cancer. J Gastrointest Surg 2002;6:838–843.
Sato N, Fukushima N, Maehara N, et al. SPARC/osteonectin is a frequent target for aberrant methylation in pancreatic adenocarcinoma and a mediator of tumor–stromal interactions. Oncogene 2003;22:5021–5030.
Maitra A, Iacobuzio-Donahue C, Rahman A, et al. Immunohistochemical validation of a novel epithelial and a novel stromal marker of pancreatic ductal adenocarcinoma identified by global expression microarrays: sea urchin fascin homolog and heat shock protein 47. Am J Clin Pathol 2002;118:52–59.
Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, et al. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene 2002;21:4587–4594.
Han H, Bearss DJ, Browne LW, et al. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res 2002;62:2890–2896.
Zolnierowicz S . Type 2A protein phosphatase, the complex regulator of numerous signaling pathways. Biochem Pharmacol 2000;60:1225–1235.
Ball AR, Schmiesing JA, Zhou C, et al. Identification of a chromosome-targeting domain in the human condensation subunit CNAP1/hCAP-D2/Eg7. Mol Cell Biol 2002;22:5769–5781.
Tang Q, Staub CM, Gao G, et al. A novel transforming growth factor-beta receptor-interacting protein that is also a light chain of the motor protein dynein. Mol Biol Cell 2002;13:4484–4496.
Oka T, Ouchida M, Koyama M, et al. Gene silencing of the tyrosine phosphatase SHP1 gene by aberrant methylation in leukemias/lymphomas. Cancer Res 2002;62:6390–6394.
Opdam FJ, Kamps G, Croes H, et al. Expression of Rab small GTPases in epithelial Caco-2 cells: Rab21 is an apically located GTP-binding protein in polarised intestinal epithelial cells. Eur J Cell Biol 2000;79:308–316.
Acknowledgements
We thank Dr Barbara Lee Smith Pierce (Adjunct Professor, University of Maryland University College) for editorial work in the preparation of this manuscript. This work was supported by the Japanese Ministries of Education, Culture, Sports, Science and Technology, and Health, Labor and Welfare, Vehicle Racing Commemorative Foundation, and Foundation for Promotion of Cancer Research in Japan.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Yamanaka, S., Sunamura, M., Furukawa, T. et al. Chromosome 12, frequently deleted in human pancreatic cancer, may encode a tumor-suppressor gene that suppresses angiogenesis. Lab Invest 84, 1339–1351 (2004). https://doi.org/10.1038/labinvest.3700160
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700160
Keywords
This article is cited by
-
A cDNA microarray analysis identifies 52 genes associated with cis-diamminedichloroplatinum susceptibility in head and neck squamous cell carcinoma cell lines
European Archives of Oto-Rhino-Laryngology (2010)
-
Upregulation of IGF2 is associated with an acquired resistance for cis-diamminedichloroplatinum in human head and neck squamous cell carcinoma
European Archives of Oto-Rhino-Laryngology (2010)
-
The PMAIP1 Gene on Chromosome 18 is a Candidate Tumor Suppressor Gene in Human Pancreatic Cancer
Digestive Diseases and Sciences (2008)
-
AURKA is one of the downstream targets of MAPK1/ERK2 in pancreatic cancer
Oncogene (2006)