Abstract
Composite lymphomas are rare combinations of Hodgkin's lymphoma (HL) and non-Hodgkin's lymphoma in the same patient, where clonal relatedness has been observed in most of the few cases analyzed. Here, we report a composite classical HL and diffuse large B-cell lymphoma (DLBCL) with interesting molecular features. Micromanipulation of single cells and analysis of V gene rearrangements revealed clonal relatedness with shared and distinct mutations, indicative of derivation from a common germinal center (GC) B-cell precursor and also of further development of both lymphomas in a GC. In the DLBCL, a very high mutation load, including inactivating mutations, and two copies of the same clonal rearrangement with different mutations in single cells were observed. Intriguingly, in the DLBCL precursor somatic hypermutation activity continued after acquisition of destructive V gene mutations, a feature previously found only in Epstein–Barr virus (EBV) infected B-cell expansions. Furthermore, we found evidence of light-chain receptor revision in the lymphoma precursor during a GC reaction. Re-expression of the V(D)J recombination machinery may enhance genomic instability in GC B cells and contribute to lymphomagenesis.
Similar content being viewed by others
Main
Traditionally, malignant lymphomas have been divided into two distinct groups, Hodgkin's lymphomas (HL) and non-Hodgkin's lymphomas (NHL). The majority of NHL have been shown to derive from either B or T cells,1 whereas the origin of the tumor cells in HL has remained unknown for many years. However, using micromanipulation of single cells and molecular analysis of the immunoglobulin (Ig) genes in HL, several reports revealed that the Hodgkin's and Reed–Sternberg (HRS) cells in most cases represent clonal populations of transformed B cells.2, 3, 4 In rare patients, occurrence of both NHL and HL is observed, either sequentially or simultaneously, and it is an interesting issue whether such composite lymphomas are clonally related or if occurrence is only coincidental or therapy-related, especially if there is a long time span between the diagnoses.
Each mature B cell carries rearranged V genes encoding the heavy and light chains of the B-cell receptor (BCR).5, 6 During somatic recombination of the V, (D), and J segments, extensive junctional diversity is created, making V gene rearrangements ideal ‘fingerprints’ for clonality analyses.7 Furthermore, as V gene rearrangements are modified in germinal center (GC) reactions by somatic hypermutation, analysis of somatic mutation patterns allows assignment of B-cell lymphomas to various maturation stages of B cells.6, 8, 9 The recent application of such analyses to a few cases of composite lymphomas showed that in most instances common lymphoma precursors existed.10, 11, 12, 13, 14, 15 For example, composite lymphomas consisting of follicular lymphoma and HL, or diffuse large B-cell lymphoma (DLBCL) and HL were reported to have clonally related V gene rearrangements.10, 12 Interestingly, the Ig rearrangements in these cases showed both shared mutations and mutations that were present only in the tumor cells of either of the lymphomas. This pattern of shared as well as distinct mutations in the clonally related lymphomas suggested that the common precursor of the two lymphomas was a GC B cell.
In this study, we analyzed a composite lymphoma with a combination of DLBCL and HL in the same lymph node. A common derivation was shown since clonally related Ig gene rearrangements were amplified from single DLBCL and HRS cells, but the Ig gene rearrangements were further diversified in the distinct lymphomas during tumor development.
Materials and methods
Patient Material
A 74-year-old woman was referred to Department of Internal Medicine, Verona University, for abdominal pain in May 2002. A computed tomography scan revealed a solid retroperitoneal mass (9 × 9 × 12 cm) infiltrating aorta, inferior vena cava and psoas muscle. Numerous enlarged abdominal lymph nodes were also detected. A fine-needle aspiration of the mass showed a cytology consistent with a DLBCL with frequent multilobated morphology. A large lymph node was palpable in inguinal right region and surgically removed; no other superficial lymph nodes were detected. Histological examination of the inguinal lymph node biopsy revealed a composite lymphoma with DLBCL and classical HL of mixed cellularity type. A bilateral bone marrow biopsy was additionally performed, showing involvement of HL, wheras no evidence of DLBCL infiltration was found. The patient was considered to be in stage IVB. After six cycles of rituximab–CHOP (cyclophosphamide, doxorubicine, vincristine, prednisone), the patient responded well with subtotal regression of the lymphoid masses and good peripheral blood count. At 12 months from diagnosis, the patient's general clinical condition appeared good.
Immunohistochemistry, EBER In Situ Hybridization and Micromanipulation
Sections of formalin-fixed, paraffin-embedded material were deparaffinized and stained with antibodies for CD30, CD15, CD20, BOB1, and Oct2 using standard procedures. In situ hybridization analysis for EBV-encoded RNA (EBER) was performed as previously described.16 For micromanipulation, 5-μm-thick frozen tissue sections were stained with anti-CD20 (L26, Dako, Hamburg, Germany) or anti-CD30 monoclonal antibodies (BerH2, Dako) and visualized using the avidin–biotin complex (ABC) technique. Single CD30+ HRS cells were micromanipulated with a hydraulic micromanipulator as described earlier.17 For CD20+ DLBCL cells, single cells as well as groups of cells (2–4 cells) were micromanipulated. Isolated cells were stored at −20°C in 20 μl of 1 × Expand buffer without MgCl2 (Roche Diagnostics, Mannheim, Germany). For each set of 10 micromanipulated cells or set of 10 clusters of cells, four aliquots of buffer covering the sections were aspirated and used as negative controls in the PCR.
Single-cell PCR Analysis
Single cells or groups of cells were digested with 0.5 mg/ml proteinase K (Roche Diagnostics, Mannheim, Germany) in 1 × Expand buffer for 2 h at 50°C, which was followed by enzyme inactivation at 95°C for 10 min. Amplification of the rearranged Ig genes was performed in a seminested approach, using family-specific VH leader primers or framework region (FR)I family-specific VH, Vκ and Vλ primers together with two sets of J primers designed for each locus as previously described.18, 19, 20 Amplification of DH–JH rearrangements was performed using DH family-specific primers together with JH primers as detailed earlier.21 This PCR also detects germline configuration of the IgH locus. For amplification of rearrangements between a recombination signal sequence in the Jκ–Cκ intron (Jκ–Cκ intron-RSS) region and the kappa-deleting element (KDE) and/or germline configuration of the KDE, one JκCκ primer, one Cκ primer and one first and second round KDE primer were used as described previously.22 PCR products were gel-purified and directly sequenced on an automated sequencer (ABI 3100, Applied Biosystems, Weiterstadt, Germany) using the Big Dye Terminator cycle sequencing kit. Sequences were aligned to Ig sequences from the GenBank and IMGT databases.23
Results
Histology and Immunohistology
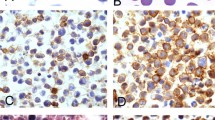
Histological examination of the inguinal lymph node, which measured 5 cm in maximal dimension, showed effacement of the normal structure by two distinct patterns of infiltration. In one area, there was a typical mixed cellular infiltrate of mixed cellularity HL with few HRS cells intermingled. In the same lymph node, separated from the HL, a more homogeneous infiltrate of large-sized B-cell blasts of a DLBCL was observed (Figure 1). Immunohistochemistry demonstrated that the HRS cells were CD30+, CD15+/−, and CD20−, whereas the DLBCL cells were CD30−, CD15−, and CD20+ (Figure 1). Furthermore, the DLBCL cells expressed BOB1 and Oct2, whereas these transcription factors were not expressed in the HRS cells (not shown). In situ hybridization for the small noncoding RNAs of EBV revealed that neither the HRS nor the DLBCL cells were EBV-infected (not shown).
Histology and immunohistology of a composite HL and DLBCL. (a, b, c) Overviews (× 5 magnification) of the composite lymphoma, in (a) a hemalaun/eosin (HE), in (b) an anti-CD20 and in (c) an anti-CD3 immunostaining. In the three pictures, the DLBCL is in the upper part and the lower right part, whereas the HL is in the lower left part; (d) × 200 magnification of a HE staining from the HL part. Two HRS cells are marked by arrows; (e) × 200 magnification from a HE staining of the DLBCL; (f) anti-CD30 staining of an HRS cell (× 400); (g) anti-CD15 staining of an HRS cell (× 400); (h) anti-CD20 immunostaining of the DLBCL part (× 200); and (i) of the HL part with a CD20-negative HRS cell (× 400).
Sequence Analysis of Ig Gene Rearrangements in DLBCL and HRS Cells
Single CD30+ HRS cells and groups of 2–4 CD20+ DLBCL cells were isolated from frozen tissue sections of the composite lymphoma using micromanipulation. Cells were then subjected to seminested PCR for VH, Vκ and Vλ gene rearrangements using FRI primers and different sets of J gene segment primers. A clonal VH3–23/JH6 rearrangement (the DH gene could not be identified) was amplified from 14 of 20 CD30+ cells, whereas no VH gene rearrangement was obtained from any of the 20 samples with CD20+ cells (Table 1). The same clonal Vκ1D–39/Jκ2 rearrangement was amplified from the DLBCL cells (10 of 20 samples) and the single HRS cells (six of 20 cells). Additionally, a second Vκ/Jκ rearrangement (Vκ1D–6/Jκ2) was obtained from 14 of 20 CD30+ cells, whereas none of the CD20+ cells displayed this rearrangement (Table 1). Only two Vλ/Jλ PCR products were amplified, one from a CD30+ cell and one from a CD20+ cell, indicating that the lymphoma cells do not carry clonal Vλ rearrangements (Table 1).
Amplification was performed on a second set of isolated HRS and DLBCL cells using VH leader region primers and JH primers, which then revealed presence of the clonal VH3–23/JH6 rearrangement in both cell types: eight of 16 single CD30+ cells and 14 of 20 samples with CD20+ cells harbored this rearrangement (Table 1). The clonal VH3–23 rearrangement in the DLBCL component had a large deletion within FRI encompassing the primer binding site (Figure 2a), explaining why the initial VHFRI PCR was unsuccessful for the DLBCL cells. All clonal rearrangements obtained from HRS and DLBCL cells were in-frame and originally potentially functional (Table 1).
Nucleotide sequences of the VH3–23 gene rearrangement (a) and the Vκ1D–39 gene rearrangement (b) amplified from the DLBCL (sequence variants I and II) and HRS cells. Sequences were aligned to the most homologous germline genes. The coding sequence is shown in triplets. Dots indicate sequence homology and dashes, missing base pairs. The intron region as well as the CDRs are indicated. Shared mutations and the FRI VH3 primer binding site are highlighted. Stop codons are marked with asterisks. HL, Hodgkin's lymphoma; DLBCL, diffuse large B-cell lymphoma.
To analyze if the second IgH allele of the DLBCL and HRS cell clones contained DH/JH joints or were in germline configuration, we performed PCR with family-specific DH primers together with JH primers. For both parts of the composite lymphoma, only PCR products indicative of germline configuration of the second IgH allele were obtained (Table 1).
Analysis of Somatic Hypermutation Pattern in Rearranged V Genes
The VH and Vκ rearrangements of the DLBCL and HRS cells were all somatically mutated, but the mutation frequencies were higher in DLBCL than HRS cells: 27–28 vs 7% for the VH3–23/JH6 rearrangement and 18 vs 7% for the Vκ1D–39/Jκ2 joint. The VH and Vκ rearrangements showed mutations shared by the DLBCL and HRS cells (13 VH and 14 Vκ gene mutations in common) as well as a substantial number of mutations present only in the DLBCL or HRS cell clones (86 and 18 distinct mutations in the VH3–23/JH6 rearrangement in DLBCL and HRS cells, respectively, and 33 and eight distinct mutations in the Vκ1D–39/Jκ2 rearrangement in DLBCL and HRS cells, respectively) (Figures 2 and 3).
Genealogical tree of the clonal development in this composite lymphoma. Two hypothetical common ancestors (I and II) have been indicated. The number of shared or additional mutations in the different clones is given. KDE, kappa-deleting element; KDE unrearr., germline configuration of the KDE; JκKDE rearr., Jκ–Cκ intron–RSS/KDE rearrangement.
Assignment of the amplified sequences to germline V genes was unequivocal for the HRS cells (VH3–23 rearrangement: 31 mutations compared to VH3–23 but 43 mutations compared to VH3–66, the next homologous germline gene; Vκ1D–39: 22 mutations compared to Vκ1D–39 but 36 mutations compared to Vκ1–6 and Vκ1–16, the next homologous germline genes) and also for the DLBCL Vκ1D–39 rearrangement (49 mutations compared to Vκ1D–39 but at least 56 and 57 mutations compared to Vκ1–6 and Vκ1–16, the next homologous germline genes). The two variants (see below) of the VH rearrangement amplified from the DLBCL had the same number of differences compared to VH3–23 and VH3–53 (variant I, 101 differences) and VH3–23 and VH3–66 (variant II, 105 differences) germline V genes. However, the clonal relatedness of the HRS and DLBCL cells, based on similarities of the VH CDRIIIs and Vκ1D–39 rearrangements, together with the unequivocal assignment of the HRS cell VH rearrangement to VH3–23 and several shared mutations between the HRS and DLBCL VH rearrangements, strongly argue that also both DLBCL VH variants use the VH3–23 germline gene.
In contrast to the HRS cells, the VH gene rearrangement of the DLBCL cells carried several inactivating mutations in the VH gene: two stop codons and one large deletion of 56 bp (Figure 2). The Vκ1D–6 rearrangement, which was amplified only from HRS cells, was also somatically mutated, with a mutation frequency of 8%.
The rearrangements of the HRS cells did not show any intraclonal diversity among the 36 cells analyzed. However, sequence analysis of the VH and Vκ rearrangements of the DLBCL cell samples revealed ‘double peaks’ in the electropherograms, indicating mixed sequences, on eight positions for the VH gene rearrangement and three positions for the Vκ gene rearrangement (Figure 2). Since 2–4 DLBCL cells were analyzed together in single tubes, the sequence variations could be due to presence of two DLBCL subclones with distinct mutation patterns, or to presence of different mutated copies of the V gene rearrangements within single cells. To clarify this issue, we performed IgH analysis on 42 isolated single DLBCL cells (Table 1). From 13 cells, sequences without ‘double peaks’ were obtained, representing two distinct mutation patterns: five of these sequences showed the same two additional mutations in addition to the mutations shared by all DLBCL cells (sequence type DLBCL I), and eight cells carried six additional mutations (DLBCL II) (Figure 2a). Importantly, from another five single cells a mixture of both sequence variants was amplified. In addition, one cell showed a ‘double peak’ at a unique position. The amplification of mixed sequences from a considerable fraction of single cells indicates that the DLBCL cells harbor two differentially mutated VH3–23 rearrangements (that mixed sequences were not obtained from all cells is most likely due to the fact that the PCR efficiency for single micromanipulated cells is usually below 50%).18 Moreover, there is no significant ongoing mutation within the DLBCL clone.
Peripheral Receptor Revision in the Composite DLBCL/HL
Since we amplified one clonal Vκ/Jκ joint from both types of tumor cells of the composite lymphoma whereas a second Vκ/Jκ joint could be amplified only from the HRS cells, we further characterized the Igκ loci using a PCR to detect Jκ–Cκ intron–RSS/KDE rearrangements and also KDE germline configuration (Figure 4).22 The KDE is located 3′ of the Cκ gene, and by rearrangement of the KDE to an unrearranged Vκ gene segment or to an RSS site in the Jκ–Cκ intron, the intervening region including the Cκ gene and both κ enhancers is deleted.24, 25 Such rearrangements are mainly used to inactivate the Igκ loci in lambda-expressing cells. The PCR for KDE rearrangements further substantiates that there are likely different rearrangements at the Igκ loci in both entities of the composite lymphoma: from the HRS cells a clonal Jκ–Cκ intron-RSS/KDE rearrangement was amplified (11 of 16 cells) in addition to a PCR product indicative of germline configuration of the KDE in the second Igκ allele (10 of 16 cells), while from the DLBCL cells only PCR products showing germline configuration were obtained (18 of 20 cells) (Table 1). We could not determine if the germline KDE PCR products from the DLBCL were derived from two or only one Igκ allele, since we did not detect any polymorphisms in the region amplified with the KDE primers using DNA from whole tissue sections.
Amplification of rearrangements involving the KDE. The upper part of the scheme shows an Igκ locus with a Vκ/Jκ joint, the Jκ intron RSS, both κ enhancers, the κ constant region and the KDE. Positions of primers used to detect amplificates involving the KDE are indicated below. Rearrangements involving the KDE as well as the resulting PCR products are shown in the middle of the scheme.
The combinations of PCR products amplified from the two parts of the composite lymphoma—clonal Vκ1D–39/Jκ2, Vκ1D–6/Jκ2 and Jκ–Cκ intron–RSS/KDE rearrangements and KDE germline configuration from the HRS cells, but only the clonal Vκ1D–39/Jκ2 and KDE germline configuration from the DLBCL cells—can be explained at least in two ways. One hypothesis is that the precursor of both lymphomas contained only the Vκ1D–39/Jκ2 joint and, by receptor revision, the Vκ1D–6/Jκ2 joint was rearranged in the HRS precursor. Since the HRS and DLBCL clone share somatic V gene mutations, and since the Jκ–Cκ intron–RSS/KDE rearrangement abolishes somatic hypermutation on the respective Igκ allele (due to concomitant deletion of the κ enhancers that are needed for hypermutation),26 the Vκ1D–6/Jκ2 rearrangement and the Jκ–Cκ intron–RSS/KDE rearrangement likely happened after acquisition of mutations in the Vκ1D–39/Jκ2 joint, that is, in a GC reaction. Alternatively, both Vκ/Jκ joints may have been present in a common precursor of the two lymphomas, but the allele harboring the Vκ1D–6/Jκ2 joint was later lost in the DLBCL clone (eg by chromosomal deletion or Vκ/KDE rearrangement). Importantly, also for this scenario, the KDE rearrangement in the HRS cells should have happened after mutations accumulated in the Vκ/Jκ joints, that is, in the GC, as argued above. Hence, the presence of two mutated Vκ/Jκ joints, both using Jκ2, together with a Jκ–Cκ intron–RSS/KDE rearrangement, clearly argues that Igκ rearrangement(s) occurred during a GC reaction in the HL precursors.
Discussion
Several composite lymphomas of HL and NHL have recently been investigated using molecular techniques and a clonal relation has been found in most of these cases.10, 11, 12, 13, 14, 15 In this study, we investigated a composite lymphoma of HL and DLBCL and were able to show clonally related VH and Vκ gene rearrangements in both lymphomas, which reveals derivation from a common lymphoma precursor also in this case. The amplified Ig gene rearrangements were somatically mutated in both lymphomas, showing shared as well as distinct mutations. This combination of shared as well as distinct mutations strongly suggests that the two lymphomas derive from two distinct members of a common GC B-cell clone. Presumably, the common shared precursor already carried some transforming event(s), and further distinct genetic lesions were acquired by the precursors of the DLBCL and HL, likely also in the GC microenvironment (Figure 3).
Analyses of Ig rearrangements in B-cell lymphomas have revealed specific features for several entities and were instrumental to assign the lymphomas to various maturation stages of normal B cells.7, 8, 9 Thus, HRS cells of classical HL are considered to derive from preapoptotic GC B cells with mutated V gene rearrangements and obviously crippling mutations in a considerable fraction of cases but without ongoing somatic hypermutation,3 while DLBCL usually carry functional V gene rearrangements with or without intraclonal sequence diversity.27, 28 In the present case, intraclonal diversity was neither observed in the HRS nor in the DLBCL cells. However, the DLBCL cells carried two copies of the VH rearrangement with a distinct mutation pattern (Figure 2). This finding is most likely due to duplication of the IgH locus carrying the rearrangement and continued hypermutation after the duplication event. A similar observation was previously made in a single-cell analysis of T-cell-rich B-cell lymphomas, a subtype of DLBCL, and presence of duplications was there confirmed by FISH analysis.20 The DLBCL in the present case carried a much higher mutation load than the HRS cells, indicating that the hypermutation process was active for a longer time and more rounds of mutation in the DLBCL than in the HRS cell precursor before it was finally silenced.
Somewhat surprisingly, the DLBCL contained several crippling mutations in the VH rearrangement, whereas the HRS cells carried no obviously destructive mutations. As the second IgH alleles are in germline configuration in the lymphomas, the DLBCL could not express a functional BCR. While inactivating mutations are frequently found in HRS cells of classical HL, they are relatively rare in B-NHL.7, 29 Expression of a BCR is mandatory for normal B cell survival, and presumably also generates important survival signals for most B-cell lymphomas.8, 30 Besides classical HL, BCR-deficient B-cell clones have been found mainly in post-transplant lymphomas and in B-cell clones expanding in T-cell lymphoma of angioimmunoblastic dysproteinemia type.31, 32, 33 In these diseases, the B-cell clones are usually EBV-infected, arguing for a role of the virus (in particular LMP2a expression) in replacing the survival signal normally mediated by the BCR. Notably, the inactivating mutations in the VH rearrangement of the DLBCL (see above) were shared between both sequence variants. Consequently, the distinct mutations must have been acquired after loss of the capability to express a functional BCR, showing that hypermutation activity continued for some time after acquisition of the crippling mutations, before it was silenced.
An intriguing feature observed in this composite lymphoma are the indications for peripheral light-chain revision during lymphoma development. Light-chain replacement has been shown to take place in early B-cell development in order to rescue B cells with autoreactive properties from apoptosis by acquisition of a new VL gene rearrangement.34, 35 Over the last years, several reports indicated that receptor revision may also occur in peripheral lymphoid organs in GC reactions.36, 37, 38, 39, 40, 41, 42, 43, 44 The maturation stage of the revising cells remains, however, controversial,45, 46, 47, 48, 49, 50, 51 and an analysis of single peripheral B cells of healthy humans indicated that receptor revision in GC reactions might not contribute significantly to the diversification of the memory B-cell pool.22 A few reports observed traces for receptor revision also in B-cell malignancies, either involving replacement of VH genes or novel rearrangement in the light chain loci.13, 20, 52 In our case, we observed a configuration of the Igκ loci which, together with the mutation pattern in the Vκ/Jκ joints, indicates that light chain revision occurred during a GC reaction in a precursor of the HRS cells. If the V gene recombination machinery can indeed be reactivated in GC B cells, mistargeting of this process may sometimes contribute to lymphoma pathogenesis, for example, by causing chromosomal translocation.53
Taken together, molecular investigation of this composite lymphoma of HL and DLBCL suggests a common GC clone derivation as has been shown previously for other combinations of HL and B-cell NHL. However, in this composite lymphoma the NHL carried a crippled VH rearrangement and sustained somatic hypermutation occurred in an already BCR-less precursor. In addition, the indications for light-chain receptor revision in a GC reaction imply that activation of the V(D)J recombination machinery may enhance genomic instability in GC and also contribute to lymphomagenesis.
Accession codes
References
Jaffe ES, Harris NL, Stein H, et al. (eds). World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press: Lyon, France, 2001.
Küppers R, Rajewsky K, Zhao M, et al. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci USA 1994;91:10962–10966.
Kanzler H, Küppers R, Hansmann ML, et al. Hodgkin and Reed–Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med 1996;184:1495–1505.
Marafioti T, Hummel M, Foss HD, et al. Hodgkin and Reed–Sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood 2000;95:1443–1450.
Tonegawa S . Somatic generation of antibody diversity. Nature 1983;302:575–581.
Rajewsky K . Clonal selection and learning in the antibody system. Nature 1996;381:751–758.
Klein U, Goossens T, Fischer M, et al. Somatic hypermutation in normal and transformed human B cells. Immunol Rev 1998;162:261–280.
Küppers R, Klein U, Hansmann ML, et al. Cellular origin of human B-cell lymphomas. N Engl J Med 1999;341:1520–1529.
Stevenson FK, Sahota SS, Ottensmeier CH, et al. The occurrence and significance of V gene mutations in B cell-derived human malignancy. Adv Cancer Res 2001;83:81–116.
Bräuninger A, Hansmann ML, Strickler JG, et al. Identification of common germinal-center B-cell precursors in two patients with both Hodgkin's disease and non-Hodgkin's lymphoma. N Engl J Med 1999;340:1239–1247.
Marafioti T, Hummel M, Anagnostopoulos I, et al. Classical Hodgkin's disease and follicular lymphoma originating from the same germinal center B cell. J Clin Oncol 1999;17:3804–3809.
Küppers R, Sousa AB, Baur AS, et al. Common germinal-center B-cell origin of the malignant cells in two composite lymphomas, involving classical Hodgkin's disease and either follicular lymphoma or B-CLL. Mol Med 2001;7:285–292.
Bellan C, Lazzi S, Zazzi M, et al. Immunoglobulin gene rearrangement analysis in composite Hodgkin disease and large B-cell lymphoma: evidence for receptor revision of immunoglobulin heavy chain variable region genes in Hodgkin–Reed–Sternberg cells? Diagn Mol Pathol 2002;11:2–8.
Ohno T, Huang JZ, Wu G, et al. The tumor cells in nodular lymphocyte-predominant Hodgkin disease are clonally related to the large cell lymphoma occurring in the same individual. Direct demonstration by single cell analysis. Am J Clin Pathol 2001;116:506–511.
van den Berg A, Maggio E, Rust R, et al. Clonal relation in a case of CLL, ALCL, and Hodgkin composite lymphoma. Blood 2002;100:1425–1429.
Niedobitek G, Herbst H, Young LS, et al. Patterns of Epstein–Barr virus infection in non-neoplastic lymphoid tissue. Blood 1992;79:2520–2526.
Küppers R, Hansmann ML, Rajewsky K . Micromanipulation and PCR analysis of single cells from tissue sections. In: Herzenberg LA, Herzenberg LA, Weir MD, Blackwell C (eds). Weir's Handbook of Experimental Immunology. Blackwell Science: Malden, MA, 1997, pp 206.1–206.4.
Küppers R, Zhao M, Hansmann ML, et al. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J 1993;12:4955–4967.
Braeuninger A, Küppers R, Strickler JG, et al. Hodgkin and Reed–Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells. Proc Natl Acad Sci USA 1997;94:9337–9342.
Bräuninger A, Küppers R, Spieker T, et al. Molecular analysis of single B cells from T-cell-rich B-cell lymphoma shows the derivation of the tumor cells from mutating germinal center B cells and exemplifies means by which immunoglobulin genes are modified in germinal center B cells. Blood 1999;93:2679–2687.
Müschen M, Rajewsky K, Bräuninger A, et al. Rare occurrence of classical Hodgkin's disease as a T cell lymphoma. J Exp Med 2000;191:387–394.
Goossens T, Bräuninger A, Klein U, et al. Receptor revision plays no major role in shaping the receptor repertoire of human memory B cells after the onset of somatic hypermutation. Eur J Immunol 2001;31:3638–3648.
Lefranc MP, Giudicelli V, Ginestoux C, et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res 1999;27:209–212.
Klobeck HG, Zachau HG . The human CK gene segment and the kappa deleting element are closely linked. Nucleic Acids Res 1986;14:4591–4603.
Siminovitch KA, Bakhshi A, Goldman P, et al. A uniform deleting element mediates the loss of kappa genes in human B cells. Nature 1985;316:260–262.
Betz AG, Milstein C, Gonzalez-Fernandez A, et al. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell 1994;77:239–248.
Küppers R, Rajewsky K, Hansmann ML . Diffuse large cell lymphomas are derived from mature B cells carrying V region genes with a high load of somatic mutation and evidence of selection for antibody expression. Eur J Immunol 1997;27:1398–1405.
Lossos IS, Okada CY, Tibshirani R, et al. Molecular analysis of immunoglobulin genes in diffuse large B-cell lymphomas. Blood 2000;95:1797–1803.
Küppers R . Molecular biology of Hodgkin's lymphoma. Adv Cancer Res 2002;84:277–312.
Lam KP, Kühn R, Rajewsky K . In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell 1997;90:1073–1083.
Bräuninger A, Spieker T, Willenbrock K, et al. Survival and clonal expansion of mutating ‘forbidden’ (immunoglobulin receptor-deficient) Epstein–Barr virus-infected B cells in angioimmunoblastic T cell lymphoma. J Exp Med 2001;194:927–940.
Timms JM, Bell A, Flavell JR, et al. Target cells of Epstein–Barr-virus (EBV)-positive post-transplant lymphoproliferative disease: similarities to EBV-positive Hodgkin's lymphoma. Lancet 2003;361:217–223.
Bräuninger A, Spieker T, Mottok A, et al. Epstein–Barr virus (EBV)-positive lymphoproliferations in post-transplant patients show immunoglobulin V gene mutation patterns suggesting interference of EBV with normal B cell differentiation processes. Eur J Immunol 2003;33:1593–1602.
Nussenzweig MC . Immune receptor editing: revise and select. Cell 1998;95:875–878.
Nemazee D . Receptor selection in B and T lymphocytes. Annu Rev Immunol 2000;18:19–51.
Han S, Dillon SR, Zheng B, et al. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science 1997;278:301–305.
Papavasiliou F, Casellas R, Suh H, et al. V(D)J recombination in mature B cells: a mechanism for altering antibody responses. Science 1997;278:298–301.
de Wildt RM, Hoet RM, van Venrooij WJ, et al. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol 1999;285:895–901.
Hertz M, Kouskoff V, Nakamura T, et al. V(D)J recombinase induction in splenic B lymphocytes is inhibited by antigen-receptor signalling. Nature 1998;394:292–295.
Meffre E, Papavasiliou F, Cohen P, et al. Antigen receptor engagement turns off the V(D)J recombination machinery in human tonsil B cells. J Exp Med 1998;188:765–772.
Brard F, Shannon M, Prak EL, et al. Somatic mutation and light chain rearrangement generate autoimmunity in anti-single-stranded DNA transgenic MRL/lpr mice. J Exp Med 1999;190:691–704.
Wilson PC, Wilson K, Liu YJ, et al. Receptor revision of immunoglobulin heavy chain variable region genes in normal human B lymphocytes. J Exp Med 2000;191:1881–1894.
Hansen A, Dörner T, Lipsky PE . Use of immunoglobulin variable-region genes by normal subjects and patients with systemic lupus erythematosus. Int Arch Allergy Immunol 2000;123:36–45.
Itoh K, Meffre E, Albesiano E, et al. Immunoglobulin heavy chain variable region gene replacement as a mechanism for receptor revision in rheumatoid arthritis synovial tissue B lymphocytes. J Exp Med 2000;192:1151–1164.
Gartner F, Alt FW, Monroe RJ, et al. Antigen-independent appearance of recombination activating gene (RAG)-positive bone marrow B cells in the spleens of immunized mice. J Exp Med 2000;192:1745–1754.
Yu W, Nagaoka H, Jankovic M, et al. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature 1999;400:682–687.
Kuwata N, Igarashi H, Ohmura T, et al. Cutting edge: absence of expression of RAG1 in peritoneal B-1 cells detected by knocking into RAG1 locus with green fluorescent protein gene. J Immunol 1999;163:6355–6359.
Igarashi H, Kuwata N, Kiyota K, et al. Localization of recombination activating gene 1/green fluorescent protein (RAG1/GFP) expression in secondary lymphoid organs after immunization with T-dependent antigens in rag1/gfp knockin mice. Blood 2001;97:2680–2687.
Meffre E, Davis E, Schiff C, et al. Circulating human B cells that express surrogate light chains and edited receptors. Nat Immunol 2000;1:207–213.
Nagaoka H, Gonzalez-Aseguinolaza G, Tsuji M, et al. Immunization and infection change the number of recombination activating gene (RAG)-expressing B cells in the periphery by altering immature lymphocyte production. J Exp Med 2000;191:2113–2120.
Meru N, Jung A, Baumann I, et al. Expression of the recombination-activating genes in extrafollicular lymphocytes but no apparent reinduction in germinal center reactions in human tonsils. Blood 2002;99:531–537.
Stamatopoulos K, Kosmas C, Stavroyianni N, et al. Evidence for immunoglobulin heavy chain variable region gene replacement in a patient with B cell chronic lymphocytic leukemia. Leukemia 1996;10:1551–1556.
Küppers R, Dalla-Favera R . Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 2001;20:5580–5594.
Acknowledgements
We are grateful to Yvonne Blum and Sabine Albrecht for skilful technical assistance. This work was supported by stipends from the Swedish Society for Medical Research and the Wenner–Gren Foundations to R Rosenquist and by a Heisenberg stipend from the Deutsche Forschungsgemeinschaft to R Küppers and the Fondazione Cassa di Risparmio Verona (Bando 2001) and AIRC to F Menestrina.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rosenquist, R., Menestrina, F., Lestani, M. et al. Indications for peripheral light-chain revision and somatic hypermutation without a functional B-cell receptor in precursors of a composite diffuse large B-cell and Hodgkin's lymphoma. Lab Invest 84, 253–262 (2004). https://doi.org/10.1038/labinvest.3700025
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700025
Keywords
This article is cited by
-
Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma
Nature Communications (2019)
-
Concurrent mucosa-associated lymphoid tissue lymphoma with diffuse large B-cell lymphoma transformation and Hodgkin lymphoma of the neck
Annals of Hematology (2018)
-
Composite lymphoma in the anterior mediastinum: a case report and review of the literature
Diagnostic Pathology (2011)
-
Insights into the multistep transformation process of lymphomas: IgH-associated translocations and tumor suppressor gene mutations in clonally related composite Hodgkin's and non-Hodgkin's lymphomas
Leukemia (2005)