Abstract
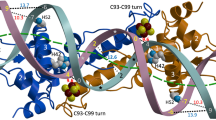
THE three-dimensional crystal structure of the Escherichia coli methionine represser, MetJ, complexed with a DNA operator fragment is described in an accompanying article1. The complex exhibits several novel features of DNA–protein interaction. DNA sequence recognition is achieved largely by hydrogen-bond contacts between the bases and amino-acid side chains located on a β-ribbon, a mode of recognition previously hypothesized on the basis of modelling of idealized β-strands and DNA2, and mutagenesis of the Salmonella phage P22 repressers Arc and Mnt3. The complex comprises a pair of MetJ represser dimers which bind to adjacent met-box sites on the DNA, and contact each other by means of a pair of antiparallel α-helices. Here we assess the importance of these contacts, and also of contacts that would be made between the C-helices of the protein, and DNA in a previous model of the complex4, by studying mutations aimed at disrupting them. The role of the carboxy-terminal helix face in operator binding was unclear, but we demonstrate that recognition of operator sequences occurs through side chains in the β-strand motif and that dimer–dimer interactions are required for effective repression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Somers, W. S. & Phillips, S. E. V. Nature 359, 387–393 (1992).
Church, G. M., Sussman, J. L., & Kim, S.-H. Proc. natn. Acad. Sci. U.S.A. 74, 1458–1462 (1977).
Knight, K. L., Bowie, J. U., Vershon, A. K., Kelly, R. D. & Sauer, R. T. J. biol. Chem. 264, 3639–3642 (1989).
Rafferty, J. B., Somers, W. S., Saint-Girons, I. & Phillips, S. E. V. Nature 341, 705–710 (1989).
Phillips, S. E. V. et al. Nature 341, 710–714 (1989).
Davidson, B. & Saint-Girons, I. Molec. Microbiol. 3, 1639–1649 (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
He, YY., McNally, T., Manfield, I. et al. Probing met represser–operator recognition in solution. Nature 359, 431–433 (1992). https://doi.org/10.1038/359431a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/359431a0
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.