Abstract
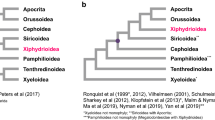
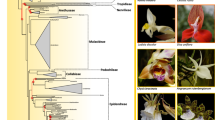
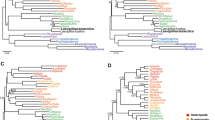
The animal phylum Arthropoda is very useful for the study of body plan evolution given its abundance of morphologically diverse species and our profound understanding of Drosophila development1. However, there is a lack of consistently resolved phylogenetic relationships between the four extant arthropod subphyla, Hexapoda, Myriapoda, Chelicerata and Crustacea. Recent molecular studies2,3,4 have strongly supported a sister group relationship between Hexapoda and Crustacea, but have not resolved the phylogenetic position of Chelicerata and Myriapoda. Here we sequence the mitochondrial genome of the centipede species Lithobius forficatus and investigate its phylogenetic information content. Molecular phylogenetic analysis of conserved regions from the arthropod mitochondrial proteome yields highly resolved and congruent trees. We also find that a sister group relationship between Myriapoda and Chelicerata is strongly supported. We propose a model to explain the apparently parallel evolution of similar head morphologies in insects and myriapods.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Akam, M. Arthropods: developmental diversity within a (super) phylum. Proc. Natl Acad. Sci. USA 97, 4438–4441 (2000).
Boore, J. L., Labrov, D. V. & Brown, W. M. Gene translocation links insects and crustaceans. Nature 392, 667–668 (1998).
Shultz, J. W. & Regier, J. C. Phylogenetic analysis of arthropods using two nuclear protein-encoding genes supports a crustacean + hexapod clade. Proc. R. Soc. Lond. B 267, 1011–1019 (2000).
Friedrich, M. & Tautz, D. Ribosomal DNA phylogeny of the major extant arthropod classes and the evolution of myriapods. Nature 376, 165–167 (1995).
Snodgrass, R. E. Evolution of Annelida, Onychophora and Arthropoda. Smithson. Misc. Coll. 138, 1–77 (1938).
Cisne, J. L. Trilobites and the evolution of arthropods. Science 186, 13–18 (1974).
Giribet, G. & Ribera, C. A review of arthropod phylogeny: new data based on ribosomal DNA sequences and direct character optimization. Cladistics 16, 204–231 (2000).
Boore, J. L., Collins, T. M., Stanton, D., Daehler, L. L. & Brown, W. M. Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature 376, 163–165 (1995).
Curole, J. P. & Kocher, T. D. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 14, 394–203 (1999).
Boore, J. L. Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780 (1999).
Wilson, K., Cahill, V., Ballment, E. & Benzie, J. The complete sequence of the mitochondrial genome of the crustacean Penaeus monodon: are malacostracan crustaceans more closely related to insects than to branchiopods? Mol. Biol. Evol. 17, 863–874 (2000).
Garcia-Machado, E. et al. Mitochondrial genes collectively suggest the paraphyly of Crustacea with respect to Insecta. J. Mol. Evol. 49, 142–149 (1999).
Aguinaldo, A. et al. Evidence for a clade of nematodes, arthropods and other molting animals. Nature 387, 489–493 (1997).
Sidow, A. & Thomas, W. K. A molecular evolutionary framework for eukaryotic model organisms. Curr. Biol. 4, 596–603 (1994).
Hausdorf, B. Early evolution of the Bilateria. Syst. Biol. 49, 130–142 (2000).
Strimmer, K. & von Haeseler, A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl Acad. Sci. USA 94, 6815–6819 (1997).
Peterson, K. J. & Eernisse, D. J. Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA sequences. Evol. Dev. 3, 170–205 (2001).
Cook, C. E., Smith, M. L., Telford, M. J., Bastianello, A. & Akam, M. Hox genes and the phylogeny of the arthropods. Curr. Biol. 11, 759–763 (2001).
Kusche, K. & Burmester, T. Diplopod hemocyanin sequence and the evolution of the Myriapoda. Mol. Biol. Evol. 18, 1566–1573 (2001).
Casares, F. & Mann, R. S. Control of antennal versus leg development in Drosophila. Nature 392, 723–726 (1998).
Delle Cave, L. & Simonetta, A. M. in The Early Evolution of Metazoa and the Significance of Problematic Taxa (eds Simonetta, A. M. & Conway Morris, S.) 189–244 (Cambridge Univ. Press, Cambridge, 1991).
Telford, M. J. & Thomas, R. H. Expression of homeobox genes shows chelicerate arthropods retain their deutocerebral segment. Proc. Natl Acad. Sci. USA 95, 10671–10675 (1998).
Damen, W. G., Hausdorf, M., Seyfarth, E. A. & Tautz, D. A conserved mode of head segmentation in arthropods revealed by the expression pattern of Hox genes in a spider. Proc. Natl Acad. Sci. USA 95, 10665–10670 (1998).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Foster, P. G. & Hickey, D. A. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol. 48, 284–290 (1999).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Muse, S. V. & Kosakovsky Pond, S. L. Hy-Phy 0.7 β (North Carolina State Univ., Raleigh, 2000).
Strimmer, K. & von Haeseler, A. Quartet puzzling—a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13, 964–969 (1996).
Adachi, J. & Hasegawa, M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J. Mol. Evol. 42, 459–468 (1996).
Felsenstein, J. PHYLIP (Phylogeny Inference Package). (Univ. Washington, Seattle, 1995).
Acknowledgements
We thank N. Muqim for technical assistance and A. Minelli and T. Burmester for comments on the manuscript. Most computation was performed on the Biological Software Server of the Institute Pasteur Paris. This study was in part supported by a DFG grant to D.T. and a Brain Korea 21 Project grant to W.K. U.W.H. was supported by fellowships from Deutscher Akademischer Austauschdienst, Korea Science and Engineering Foundation, and the Brain Korea 21 Project (Medical Sciences, Yonsei University).
Author information
Authors and Affiliations
Corresponding authors
Supplementary information

Supplement Figure 1
(GIF 12.6 KB)
Consistent support for a chelicerate/myriapod sister clade when nematode taxa are included. Single protein alignments (23PND1.PHYLIP, 23PND2_PHYLIP, 23PND3.PHYLIP, 23PND4.PHYLIP, 23PND4l.PHYLIP, 23PND5.PHYLIP, 23PND6.PHYLIP, 23PCOI-1.PHYLIP, 23PCOII-1.PHYLIP, 23PCOIII-1.PHYLIP, 23PCytB.PHYLIP and 23Patpase6-1.PHYLIP) were generated for same set of species included in the original analysis plus the four nematode species and deposited with supplementary materials. Mitochondrial protein quartet puzzling trees shown as unrooted phylograms of were estimated with a 2055 site long alignment (23P2055, EBI WWW server accession number ALIGN_000113) produced with the Gblocks program at default settings as described in Material and Methods. Bars represent 0.1 substitutions per site. a, Taxon choice as with 18P2560 and 18P1528. Myriapod/chelicerate clade supported with BP = 95. b, Taxon choice same as in a with four nematode taxa added (Caenorhabditis, Ascaris, Onchocerca, Trichinella) and the fast evolving chelicerate Ixodes removed. All four nematode taxa form extremely long branches consistent with accelerated substitution rates indicated in distance matrix (Supplement Table 1). Myriapod/chelicerate clade supported with BP = 98. c, Taxon choice same as in b with fast evolving chelicerate taxa added (Rhipicephalus, Ixodes). The latter are drawn away from the chelicerate Limulus most likely due to long branch attraction. Myriapod (Lithobius) /chelicerate (Limulus) clade supported with BP = 88.
Rights and permissions
About this article
Cite this article
Hwang, U., Friedrich, M., Tautz, D. et al. Mitochondrial protein phylogeny joins myriapods with chelicerates. Nature 413, 154–157 (2001). https://doi.org/10.1038/35093090
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35093090
This article is cited by
-
The complete mitochondrial genome of Wellcomia compar (Spirurina: Oxyuridae) and its genome characterization and phylogenetic analysis
Scientific Reports (2023)
-
Evolutionary biogeography of the centipede genus Ethmostigmus from Peninsular India: testing an ancient vicariance hypothesis for Old World tropical diversity
BMC Evolutionary Biology (2019)
-
Evolutionary origin of type IV classical cadherins in arthropods
BMC Evolutionary Biology (2017)
-
Inferring explicit weighted consensus networks to represent alternative evolutionary histories
BMC Evolutionary Biology (2013)
-
Arthropod fossil data increase congruence of morphological and molecular phylogenies
Nature Communications (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.