Abstract
Exit from mitosis requires the inactivation of mitotic cyclin-dependent kinases (CDKs). In the budding yeast, Saccharomyces cerevisiae, inactivation of CDKs during late mitosis involves degradation of B-type cyclins as well as direct inhibition of cyclin–CDK complexes by the CDK-inhibitor protein Sic1 (refs 1,2,3). Several striking similarities exist between Sic1 and Cdc6, a DNA replication factor essential for the formation of pre-replicative complexes at origins of DNA replication4,5,6,7,8,9. Transcription of both genes is activated during late mitosis by a process dependent on Swi5 (ref. 10). Like Sic1, Cdc6 binds CDK complexes in vivo11,12 and downregulates them in vitro11. Here we show that Cdc6, like Sic1, also contributes to inactivation of CDKs during late mitosis in S. cerevisiae. Deletion of the CDK-interacting domain of Cdc6 does not inhibit the function of origins of DNA replication during S phase, but instead causes a delay in mitotic exit; this delay is accentuated in the absence of Sic1 or of cyclin degradation. By contributing to mitotic exit and inactivation of CDKs, Cdc6 helps to create the conditions that are required for its subsequent role in the formation of pre-replicative complexes at origins of DNA replication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Donovan, J. D., Toyn, J. H., Johnson, A. L. & Johnston, L. H. p40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 8, 1640–1653 (1994).
Schwab, M., Schulze-Lutum, A. & Seufert, W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90, 683–693 (1997).
Visintin, R., Prinz, S. & Amon, A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 17, 460–463 (1997).
Liang, C., Weinreich, M. & Stillman, B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81, 667–676 (1995).
Cocker, J. H., Piatti, S., Santocanale, C., Nasmyth, K. & Diffley, J. F. X. An essential role for the Cdc6p protein in budding yeast pre-replicative complexes. Nature 379, 180–182 (1996).
Aparicio, O. M., Weinstein, D. M. & Bell, S. P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69 (1997).
Detweiler, C. S. & Li, J. Cdc6p establishes and maintains a state of replication competence during G1 phase. J. Cell Sci. 110, 753–763 (1997).
Donovan, S., Harwood, J., Drury, L. S. & Diffley, J. F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA 94, 5611–5616 (1997).
Tanaka, T., Knapp, D. & Nasmyth, K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90, 649–660 (1997).
Spellman, P. T. et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273–3297 (1998).
Elsasser, S., Lou, F., Wang, B., Campbell, J. L. & Jong, A. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol. Biol. Cell 7, 1723–1735 (1996).
Calzada, A., Sánchez, M., Sánchez, E. & Bueno, A. The stability of the Cdc6 protein is regulated by CDK/cyclin B complexes in Saccharomyces cerevisiae. J. Biol. Chem. 275, 9734–9741 (2000).
Drury, L. S., Perkins, G. & Diffley, J. F. X. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 16, 5966–5976 (1997).
Yeong, F. M., Lim, H. H., Padmashree, C. G. & Surana, U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Mol. Cell 5, 501–511 (2000).
Irniger, S., Piatti, S., Michaelis, C. & Nasmyth, K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81, 269–278 (1995).
King, R. W. et al. 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81, 279–288 (1995).
Peters, J. M. Subunits and substrates of the anaphase-promoting complex. Exp. Cell Res. 248, 339–349 (1999).
Pichler, S., Piatti, S. & Nasmyth, K. Is the yeast Anaphase Promoting Complex needed to prevent re-replication during G2 and M phases? EMBO J. 16, 5988–5997 (1997).
Yuste-Rojas, M. & Cross, F. R. Mutations in CDC14 result in high sensitivity to cyclin gene dosage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263, 60–72 (2000).
Visintin, R. et al. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2, 709–718 (1998).
Culotti, J. & Hartwell, L. H. Genetic control of the cell division cycle in yeast III. Seven genes controlling nuclear division. Exp. Cell Res. 67, 389–401 (1971).
Mizushima, T., Takahashi, N. & Stillman, B. Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes Dev. 14, 1631–1641 (2000).
Zwerschke, W., Rottjakob, H.-W. & Kuntzel, H. The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes an ATP/GTPase controlling S phase initiation. J. Biol. Chem. 269, 23352–23356 (1994).
Piatti, S., Lengauer, C. & Nasmyth, K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14, 3788–3799 (1995).
Hogan, E. & Koshland, D. Addition of extra origins of replication to a minichromosome suppressed its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 89, 3098–3102 (1992).
Bähler, J. & Pringle, J. R. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 12, 1356–1370 (1998).
Acknowledgements
We thank J. Correa, K. Labib, S. Moreno, L. Pelloquin and the ASP laboratory for many discussions, and K. Labib for critically reading this manuscript. We are grateful to R. Basco, J. Correa, J. Diffley, S. Ufano and C. Vázquez for yeast strains and anti-Pgk antibody. We also thank N. Skinner for editing the text. This research was supported by CICYT and PGC grants to A.B.
Author information
Authors and Affiliations
Corresponding author
Supplementary information

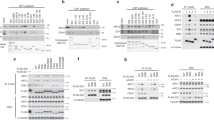
Figure 1
(JPG 24.7 KB)
Activation of chromosomal DNA replication origins in D47cdc6 cells. Left and middle panels, two-dimensional gels of NcoI digested DNA from asynchronous cultures of wild-type or D47cdc6 strains hybridized with a 5 kb NcoI fragment containing ARS1. A drawing of how replication intermediates distribute on (N/N) two-dimensional gels.

Figure 2
(JPG 110 KB)
A nocodazole block-and-release experiment with wild-type, D47cdc6 and Dhct1 strains. Samples taken at the indicated intervals were processed for DNA content analysis (a), Clb2-associated H1 kinase activity (b and e, left panel), Westerns analysis (c, d and e, middle panel) and mitotic spindle analysis (e, right panel).

Figure 3
(JPG 62.2 KB)
Genetic interactions among cdc14-1 and cdc23-1 ts alleles and the D47cdc6 amino-terminal truncation. Ten-fold dilutions of wild-type, D47cdc6, cdc14-1, cdc23-1, cdc14-1 D47cdc6 and cdc23-1 D47cdc6 isogenic S. cerevisiae strains from cultures in log phase at 22°C were inoculated in YEPD Petri dishes and incubated at the indicated temperatures.

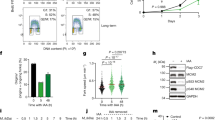
Figure 4
(JPG 25.2 KB)
SIC1 rescues the late-in-mitosis defect observed in D47cdc6 Dhct1 cells. (a), Clb2-associated H1 kinase assays and Western analysis in asynchronous D47cdc6 Dhct1 GAL1-10:SIC1 cells repressed (lane 1) or induced (lane 3) for SIC1 expression. Nocodazole samples (lanes 2 and 4) and untagged Clb2 control (C) are shown for reference. (b), Extended spindles analysis of the same strain when SIC1 is off and on.
Rights and permissions
About this article
Cite this article
Calzada, A., Sacristán, M., Sánchez, E. et al. Cdc6 cooperates with Sic1 and Hct1 to inactivate mitotic cyclin-dependent kinases. Nature 412, 355–358 (2001). https://doi.org/10.1038/35085610
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35085610
This article is cited by
-
Dynamics of ASXL1 mutation and other associated genetic alterations during disease progression in patients with primary myelodysplastic syndrome
Blood Cancer Journal (2014)
-
APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit
Nature (2002)
-
Competence to replicate in the unfertilized egg is conferred by Cdc6 during meiotic maturation
Nature (2002)
-
Origin licensing and programmed cell death: a hypothesis
Cell Death & Differentiation (2002)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.