Abstract
Programmed cell death (apoptosis) is a tightly regulated process of cell disassembly in which dying cells and their nuclei shrink and fragment and the chromosomal DNA is degraded into internucleosomal repeats1,2,3. Here we report the characterization of the cps-6 gene, which appears to function downstream of, or in parallel to, the cell-death protease CED-3 of Caenorhabditis elegans in the DNA degradation process during apoptosis. cps-6 encodes a homologue of human mitochondrial endonuclease G4,5, and its protein product similarly localizes to mitochondria in C. elegans. Reduction of cps-6 activity caused by a genetic mutation or RNA-mediated interference (RNAi) affects normal DNA degradation, as revealed by increased staining in a TUNEL assay, and results in delayed appearance of cell corpses during development in C. elegans. This observation provides in vivo evidence that the DNA degradation process is important for proper progression of apoptosis. CPS-6 is the first mitochondrial protein identified to be involved in programmed cell death in C. elegans, underscoring the conserved and important role of mitochondria in the execution of apoptosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kerr, J. F., Wyllie, A. H. & Currie, A. R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257 (1972).
Steller, H. Mechanisms and genes of cellular suicide. Science 267, 1445–1449 (1995).
Nagata, S. Apoptotic DNA fragmentation. Exp. Cell Res. 256, 12–18 (2000).
Ruiz-Carrillo, A. & Renaud, J. Endonuclease G: a (dG)n × (dC)n-specific DNase from higher eukaryotes. EMBO J. 6, 401–407 (1987).
Cote, J. & Ruiz-Carrillo, A. Primers for mitochondrial DNA replication generated by endonuclease G. Science 261, 765–769 (1993).
Horvitz, H. R. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. (Suppl.) 59, 1701–1706 (1999).
Chalfie, M. The differentiation and function of the touch receptor neurons of Caenorhabditis elegans. Prog. Brain Res. 105, 179–182 (1995).
Stanfield, G. M. & Horvitz, H. R. The ced-8 gene controls the timing of programmed cell deaths in C. elegans. Mol. Cell 5, 423–433 (2000).
Li, L. Y., Luo, X. & Wang, X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature, 412, 95–99 (2001).
Hsu, D. R., Chuang, P. T. & Meyer, B. J. DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development 121, 3323–3334 (1995).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Chen, F. et al. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science 287, 1485–1489 (2000).
Wu, Y. C., Stanfield, G. M. & Horvitz, H. R. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 14, 536–548 (2000).
Liu, X., Zou, H., Slaughter, C. & Wang, X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell 89, 175–184 (1997).
Enari, M. et al. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391, 43–50 (1998).
Sulston, J. E. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. B 275, 287–297 (1976).
Hedgecock, E. M., Sulston, J. E. & Thomson, J. N. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220, 1277–1279 (1983).
Zhang, J. et al. Resistance to DNA fragmentation and chromatin condensation in mice lacking the DNA fragmentation factor 45. Proc. Natl Acad. Sci. USA 95, 12480–12485 (1998).
McIlroy, D. et al. An auxiliary mode of apoptotic DNA fragmentation provided by phagocytes. Genes Dev. 14, 549–558 (2000).
Gross, A., McDonnell, J. M. & Korsmeyer, S. J. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 13, 1899–1911 (1999).
Liu, X., Kim, C. N., Yang, J., Jemmerson, R. & Wang, X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86, 147–157 (1996).
Susin, S. A. et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397, 441–446 (1999).
Du, C., Fang, M., Li, Y., Li, L. & Wang, X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102, 33–42 (2000).
Verhagen, A. et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing inhibitor of apoptosis (IAP) proteins. Cell 102, 43–53 (2000).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Riddle, D. L., Blumenthal, T., Meyer, B. J. & Preiss, J. R. (eds) C. elegans II (Cold Spring Harbor Laboratory, Cold Spring Harbor, 1997).
Shaham, S., Reddien, P. W., Davies, B. & Horvitz, H. R. Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics 153, 1655–1671 (1999).
Tabara, H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999).
Gu, T., Orita, S. & Han, M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol. Cell Biol. 18, 4556–4564 (1998).
Clark, S. G., Lu, X. & Horvitz, H. R. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137, 987–997 (1994).
Acknowledgements
We thank A. Coulson and the C. elegans Genome Sequencing Consortium for cosmids; A. Fire for vectors; B. Hersh and R. H. Horvitz for the protocol on using Mitotracker Red; and Y. C. Wu for discussion. We also thank R. J. McIntosh, G. Odorizzi, R. Poyton and W. Wood for comments on the manuscript. J.Z.P. is supported by a NSF Graduate Fellowship and X.W. by grants from NIH and the Welch Foundation. D.X. is supported by grants from ACS and NIH and is a recipient of the BWF Career Award and the Searle Scholar Award.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Results
Several different cDNA clones have been identified from the 5.3 kb cps-6 rescuing fragment carried by pJP5.3 (Fig. 2a in the full text). We have isolated four different cDNA clones from a C. elegans two-hybrid library by PCR amplification using 5’ primers corresponding to sequences from the two-hybrid vector and a 3’ primer corresponding to the last six amino acids of worm Endo G. All four cDNA clones are approximately 1 kb in length and contain a single ORF with 308 amino acids (Fig. 2b in the full text). We have also identified one cDNA clone (yk661a12) from the C. elegans EST database (Y. Kohara, personal communications) that contains a single ORF with 239 amino acids that is approximately 1.5 kb upstream of the worm EndoG ORF. The following results indicate that the cps-6 rescuing activity of pJP5.3 or pJP9.8 is conferred by worm EndoG but not the ORF upstream of EndoG: a 1428bp deletion in pJP9.8 (pJP9.8(DBS)), which removes most of the upstream ORF, does not affect the rescuing activity of pJP9.8, but another 1494bp deletion in pJP9.8 (pJP9.8(DXba)) that removes the first exon of worm EndoG, abolishes its rescuing activity (Fig. 2a in the full text).
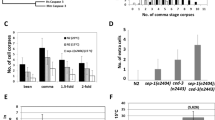
To confirm that RNAi treatment abolishes the expression of worm EndoG, we examined the expression of worm EndoG in control and EndoG RNAi-treated animals using an EndoG GFP fusion construct (Pdpy-30CPS-6::GFP). In RNAi control animals carrying the Pdpy-30CPS-6::GFP transgene, 68% of the embryos (n=128) displayed bright GFP fluorescence (Fig. 1b). In contrast, in EndoG(RNAi)-treated animals carrying the same transgene, less than 7% of the embryos (n=134) had GFP fluorescence, in most of which GFP was barely visible (Fig. 1d). These results indicate that EndoG(RNAi) treatment effectively abolishes the expression of EndoG in C. elegans.
We found that EndoG(RNAi), like the sm116 mutation, can weakly suppress ectopic neuronal deaths induced by acCED-3 or enhance the suppression of acCED-3-mediated neuronal deaths by ced-8(n1891) (Table 1).
We examined whether cps-6 genetically interacts with another nuclease-encoding gene, nuc-1. Consistent with previous studies, we found that in nuc-1(e1392) mutant, which harbors a null allele of nuc-1, the killing process of cell death and engulfment of cell corpses appear to be normal; the time-course of embryonic cell corpses of nuc-1(e1392) animals is very similar to that of wild-type animals (Fig. 2a) and there are no extra cells observed in nuc-1(e1392) animals (Table 2). When we examined the profiles of cell corpse appearance from cps-6(sm116); nuc-1(e1392) animals or cps-6(RNAi); nuc-1(e1392) animals, we found that they are very similar to that of the cps-6(sm116) or cps-6(RNAi) animals (Fig. 2b and data not shown), indicating that the timing of cell deaths is not further delayed in the double mutants. Interestingly, cps-6(sm116); nuc-1(e1392) double mutants appear to have a higher number of extra cells in the anterior pharynx than either mutant alone (an average of 0.35 extra cells; Table 2) but the number of extra cells in cps-6(RNAi); nuc-1(e1392) animals (an average of 0.29 extra cells) is not significantly higher than that of cps-6(RNAi) animals (an average of 0.2 extra cells).
Methods
Standard methods for molecular biology were used. Sequences of primers used for PCR amplification are available upon request. cps-6 cDNAs were PCR amplified from a C. elegans two-hybrid library (RB1) and subcloned into vector pBluescript KSII(+) via its Xho I and Spe I sites. One of these cDNAs was subcloned into the C. elegans expression vector pS235, which contains the dpy-30 promoter, to generate the plasmid, Pdpy-30CPS-6. Pdpy-30CPS-6::GFP was generated by inserting a 1. 85 kb Xba I-Spe I fragment from pPD95.77 which contains a GFP ORF into Pdpy-30CPS-6 via its Spe I site, which is immediately after the last amino acid of CPS-6. A full-length murine EndoG cDNA was cloned into pS235 via its Nhe I and EcoR I sites to generate the expression construct Pdpy-30mEndoG. To make the PhspCPS-6(40-308)::GFP constructs, we first cloned a cDNA fragment containing CPS-6(40-308) into the heat-shock inducible expression vectors, pPD49.78 and pPD49.83. Subsequently, an Xba I-Spe I fragment derived from pPD95.77 was cloned into pPD49.78-CPS-6(40-308) and pPD49.83-CPS-6(40-308) to generate PhspCPS-6(40-308)::GFP expression vectors.
We have attempted to identify the molecular lesion responsible for the cps-6(sm116) mutant by sequencing the entire coding region, introns, approximately 800bp upstream and 300bp downstream of the worm Endo G gene from the cps-6(sm116) mutant but did not find any mutation. Thus, sm116 may affect a cis-regulatory element important for the expression of the worm Endo G gene.
Supplementary Figures and Tables

Figure 1
(JPG 141 KB)

Figure 2
(JPG 54.5 KB)

Table 1
(JPG 56.5 KB)

Table 2
(JPG 91.3 KB)
Rights and permissions
About this article
Cite this article
Parrish, J., Li, L., Klotz, K. et al. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature 412, 90–94 (2001). https://doi.org/10.1038/35083608
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35083608
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.