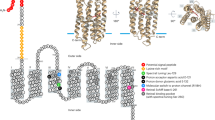
Abstract
Proteorhodopsin1, a retinal-containing integral membrane protein that functions as a light-driven proton pump, was discovered in the genome of an uncultivated marine bacterium; however, the prevalence, expression and genetic variability of this protein in native marine microbial populations remain unknown. Here we report that photoactive proteorhodopsin is present in oceanic surface waters. We also provide evidence of an extensive family of globally distributed proteorhodopsin variants. The protein pigments comprising this rhodopsin family seem to be spectrally tuned to different habitats—absorbing light at different wavelengths in accordance with light available in the environment. Together, our data suggest that proteorhodopsin-based phototrophy is a globally significant oceanic microbial process.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others

References
Béjà, O. et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902– 1906 (2000).
Oesterhelt, D. & Stoeckenius, W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature 233, 149– 152 (1971).
Henderson, R. & Unwin, P. N. Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257, 28– 32 (1975).
Danon, A. & Stoeckenius, W. Photophosphorylation in Halobacterium halobium. Proc. Natl Acad. Sci. USA 71, 1234– 1238 (1974).
Mullins, T. D., Britcshgi, T. B., Krest, R. L. & Giovannoni, S. J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40, 148– 158 (1995).
Sasaki, J. & Spudich, J. L. The transducer protein HtrII modulates the lifetimes of sensory rhodopsin II photointermediates. Biophys. J. 75, 2435– 2440 (1998).
Spudich, J. L., Yang, C. S., Jung, K. H. & Spudich, E. N. Retinylidene proteins: Structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16, 365– 392 (2000).
Neugebauer, D. C., Zingsheim, H. P. & Oesterhelt, D. Biogenesis of purple membrane in halobacteria. Methods Enzymol. 97, 218– 226 (1983).
Michel, H. & Oesterhelt, D. Electrochemical proton gradient across the cell membrane of Halobacterium halobium: effect of N,N′-dicyclohexylcarbodiimide, relation to intracellular adenosine triphosphate, adenosine diphosphate, and phosphate concentration, and influence of the potassium gradient. Biochemistry 19, 4607– 4614 (1980).
Béjà, O. et al. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2, 516– 529 (2000).
Karl, D. M. & Lukas, R. The Hawaii Ocean Time-series (HOT) program: Background, rationale and field implementation. Deep-Sea Res. 43, 129– 156 (1996).
Takahashi, T. et al. Color regulation in the archaebacterial phototaxis receptor phoborhodopsin (sensory rhodopsin II). Biochemistry 29, 8467– 8474 (1990).
Kirk, J. T. O. in Light and Photosynthesis in Aquatic Ecosystems 104– 134 (Cambridge Univ. Press, Cambridge, 1983).
Bowmaker, J. K. The ecology of visual pigments. Novartis Found. Symp. 224, 21– 31 (1999).
Urbach, E., Scanlan, D. J., Distel, D. L., Waterbury, J. B. & Chisholm, S. W. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 46, 188– 201 (1998).
Moore, L. R., Rocap, G. & Chisholm, S. W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464– 467 (1998).
Palenik, B. Chromatic adaptation in marine Synechococcus strains. Appl. Environ. Microbiol. 67, 991– 994 (2001).
Ferris, M. J. & Palenik, B. Niche adaptation in ocean cyanobacteria. Nature 396, 226– 228 (1998).
Kolber, Z. S., Van Dover, C. L., Niderman, R. A. & Falkowski, P. G. Bacterial photosynthesis in surface waters of the open ocean. Nature 407, 177– 179 (2000).
Eilers, H., Pernthaler, J., Glockner, F. O. & Amann, R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66, 3044– 3051 (2000).
Gonzalez, J. M. et al. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66, 4237– 4246 (2000).
Rappé, M. S., Vergin, K. & Giovannoni, S. J. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33, 219– 232 (2000).
DeLong, E. F. et al. Dibiphytanyl ether lipids in nonthermophilic crenarchaeotes. Appl. Environ. Microbiol. 64, 1133– 1138 (1998).
Shimono, K., Iwamoto, M., Sumi, M. & Kamo, N. Functional expression of pharaonis phoborhodopsin in Escherichia coli. FEBS Lett. 420, 54– 56 (1997).
DeLong, E. F., Taylor, L. T., Marsh, T. L. & Preston, C. M. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65, 5554– 5563 (1999).
Acknowledgements
We thank J. Zehr and D. Karl for the HOT samples and the captain and crew of the RV Point Lobos for expert assistance at sea. D. Karl and R. Letellier provided spectral irradiance data. This research was supported by the National Science Foundation (E.F.D.), the NIH (J.L.S.), and the David and Lucile Packard Foundation (E.F.D.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Béjà, O., Spudich, E., Spudich, J. et al. Proteorhodopsin phototrophy in the ocean. Nature 411, 786–789 (2001). https://doi.org/10.1038/35081051
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35081051
This article is cited by
-
Microbial circadian clocks: host-microbe interplay in diel cycles
BMC Microbiology (2023)
-
A comparative study reveals the relative importance of prokaryotic and eukaryotic proton pump rhodopsins in a subtropical marginal sea
ISME Communications (2023)
-
Rhodopsin-mediated nutrient uptake by cultivated photoheterotrophic Verrucomicrobiota
The ISME Journal (2023)
-
Pro219 is an electrostatic color determinant in the light-driven sodium pump KR2
Communications Biology (2021)
-
Marine Dadabacteria exhibit genome streamlining and phototrophy-driven niche partitioning
The ISME Journal (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.