Key Points
-
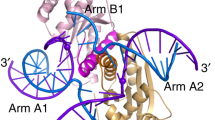
Homologous recombination is an ancient process that functions to conserve genetic integrity and create genetic diversity. It relies on the formation of Holliday junctions — four-stranded DNA structures that create physical links between DNA duplexes. This review concentrates on the endonucleases that recognize this structure and catalyse its cleavage — the junction-resolving enzymes.
-
Recent progress includes the characterization of junction-resolving enzymes in the archaea and their viruses, the identification of the pox viral resolving enzyme responsible for resolution of viral concatamers, and the detection of a possible mammalian junction migrating and resolving complex analogous to the Escherichia coli RuvABC machinery
-
Studies in E. coli have emphasized the important role of homologous recombination in rescuing stalled or collapsed DNA replication forks, and the control of crossover versus non-crossover recombinant formation by the RuvABC complex. These findings are likely to be relevant in the eukaryotes.
-
Structural and sequence analysis indicates that most of the junction-resolving enzymes can be fitted into two main superfamilies of proteins. Sequence-selective enzymes including RuvC and CceI fall into the integrase class, whereas T7 endonuclease I and Hjc are classed with nucleases. All are probably evolved from a common metal-ion-binding domain.
-
Junction-resolving enzymes bind four-way DNA junctions in dimeric form. Binding is highly structure selective, but all the enzymes significantly distort the structure of the DNA on binding. Some of the resolving enzymes also have pronounced sequence selectivity at the cleavage stage; this can effectively ensure cleavage of Holliday junctions and not other branched DNA structures.
-
Several junction-resolving enzymes show an accelerated second strand cleavage, which is particularly pronounced for RuvC. This is probably the result of a strained DNA-enzyme complex, and ensures a productive resolution event.
-
The structures of four junction-resolving enzymes have been determined by crystallography. The protein folds have little in common. The active sites of T7 endonuclease I and Hjc are very similar to those of type II restriction enzymes, suggesting a two-metal-ion cleavage mechanism.
Abstract
Junction-resolving enzymes are ubiquitous nucleases that are important for DNA repair and recombination and act on DNA molecules containing branch points, especially four-way junctions. They show a pronounced selectivity for the structure of the DNA substrate but, despite its importance, the structural selectivity is not well understood. This poses an intriguing challenge in molecular recognition on a relatively large scale.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Holliday, R. A mechanism for gene conversion in fungi. Genet. Res. 5, 282–304 (1964).
Cox, M. M. et al. The importance of repairing stalled replication forks. Nature 404, 37–41 ( 2000).
Kowalczykowski, S. C. Initiation of genetic recombination and recombination-dependent replication . Trends Biochem. Sci. 25, 156– 165 (2000).A comprehensive review of the recent advances in understanding the relationship between DNA replication and recombination.
Komori, K., Sakae, S., Shinagawa, H., Morikawa, K. & Ishino, Y. A Holliday junction resolvase from Pyrococcus furiosus : functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in bacteria, eukarya, and archaea. Proc. Natl Acad. Sci. USA 96, 8873–8878 (1999). First identification of an archaeal Holliday-junction-resolving enzyme, using random expression of Pyrococcus furiosus genes in Escherichia coli .
Kvaratskhelia, M. & White, M. F. An archaeal Holliday junction resolving enzyme from Sulfolobus solfataricus exhibits unique properties. J. Mol. Biol. 295, 193– 202 (2000).Characterization of an archaeal resolving enzyme, of probable viral origin, that cuts Holliday junctions uniquely on continuous strands.
Kvaratskhelia, M., Wardleworth, B. N., Norman, D. G. & White, M. F. A conserved nuclease domain in the archaeal Holliday junction resolving enzyme HJC. J. Biol. Chem. 275, 25540– 25546 (2000).
Kvaratskhelia, M., Wardleworth, B. N. & White, M. F. Multiple Holliday junction resolving enzyme activities in the Crenarchaeota and Euryarchaeota. FEBS Lett. 491, 243–246 ( 2001).
Stuart, D., Ellison, K., Graham, K. & McFadden, G. In vitro resolution of poxvirus replicative intermediates into linear minichromosomes with hairpin termini by a virally induced Holliday junction endonuclease. J. Virol. 66, 1551–1563 (1992).
Baroudy, B. M., Venkatesan, S. & Moss, B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell 28, 315–324 (1982).
Dickie, P., Morgan, A. R. & McFadden, G. Cruciform extrusion in plasmids bearing the replicative intermediate configuration of a poxvirus telomere. J. Mol. Biol. 196, 541–558 ( 1987).
Garcia, A. D., Aravind, L., Koonin, E. & Moss, B. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc. Natl Acad. Sci. USA 97, 8926–8931 (2001).Identification of the pox viral Holliday junction-resolving enzyme, and its relationship with RuvC and Cce1. PubMed
Lilley, D. M. J. & White, M. F. Resolving the relationships of resolving enzymes. Proc. Natl Acad. Sci. USA 97, 9351–9353 (2000).
West, S. C. & Korner, A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 82, 6445–6449 (1985).
Jeyaseelan, R. & Shanmugam, G. Human placental endonuclease cleaves Holliday junctions. Biochem. Biophys. Res. Commun. 156, 1054–1060 ( 1988).
Elborough, K. M. & West, S. C. Resolution of synthetic Holliday junctions in DNA by an endonuclease from calf thymus. EMBO J. 9, 2931–2936 ( 1990).
Hyde, H., Davies, A. A., Benson, F. E. & West, S. C. Resolution of recombination intermediates by a mammalian activity functionally analogous to Escherichia coli RuvC resolvase. J. Biol. Chem. 269, 5202–5209 ( 1994).
Constantinou, A., Davies, A. A. & West, S. C. Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell 104, 259–268 (2001). First evidence of RuvABC-like activities co-purifying in a eukaryotic cell.
Panyutin, I. G., Biswas, I. & Hsieh, P. A pivotal role for the structure of the Holliday junction in DNA branch migration. EMBO J. 14, 1819 –1826 (1995).
Duckett, D. R. et al. The structure of the Holliday junction and its resolution . Cell 55, 79–89 (1988).
Grigoriev, M. & Hsieh, P. A histone octamer blocks branch migration of a Holliday junction. Mol. Cell. Biol. 17, 7139–7150 (1997).
Rafferty, J. B. et al. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science 274, 415–421 (1996).
Hargreaves, D. et al. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6 Å resolution. Nature Struct. Biol. 5, 441–446 (1998).
Nishino, T., Ariyoshi, M., Iwasaki, H., Shinagawa, H. & Morikawa, K. Functional analyses of the domain structure in the Holliday junction binding protein RuvA. Structure 6, 11–21 (1998 ).
Roe, S. M. et al. Crystal structure of an octameric RuvA–Holliday junction complex. Mol. Cell 2, 361– 372 (1998).
Parsons, C. A., Stasiak, A., Bennett, R. J. & West, S. C. Structure of a multisubunit complex that promotes DNA branch migration. Nature 374, 375–378 ( 1995).
van Gool, A. J., Shah, R., Mézard, C. & West, S. C. Functional interactions between the Holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J. 17, 1838–1845 (1998).
Van Gool, A. J., Hajibagheri, N. M., Stasiak, A. & West, S. C. Assembly of the Escherichia coli RuvABC resolvasome directs the orientation of Holliday junction resolution. Genes Dev. 13, 1861–1870 (1999). Demonstration that the RuvAB branch migration machinery controls the choice of strands cleaved by the resolving enzyme RuvC, with implications for control of crossing over.
Cromie, G. A. & Leach, D. R. F. Control of crossing over. Mol. Cell 6, 815–826 ( 2000).A demonstration of how biases in crossover versus non-crossover recombination events in Escherichia coli are controlled by RuvABC.
Michel, B., Recchia, G. D., Penel-Colin, M., Ehrlich, S. D. & Sherratt, D. J. Resolution of Holliday junctions by RuvABC prevents dimer formation in rep mutants and UV-irradiated cells. Mol. Microbiol. 37, 180– 191 (2000).
Michel, B. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25, 173–178 ( 2000).
Postow, L. et al. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. (in the press).
McGlynn, P. & Lloyd, R. G. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression . Cell 101, 35–45 (2000).
Seigneur, M., Bidnenko, V., Ehrlich, S. D. & Michel, B. RuvAB acts at arrested replication forks. Cell 95, 419–430 (1998).
Lim, D. S. & Hasty, P. A mutation in mouse RAD51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16, 7133–7143 (1996).
Tsuzuki, T. et al. Targeted disruption of the RAD51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA 93, 6236–6240 (1996).
Ariyoshi, M. et al. Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell 78, 1063–1072 (1994).
Saito, A., Iwasaki, H., Ariyoshi, M., Morikawa, K. & Shinagawa, H. Identification of four acidic amino acids that constitute the catalytic centre of the RuvC Holliday junction resolvase. Proc. Natl Acad. Sci. USA 92, 7470–7474 (1995).
Raaijmakers, H. et al. X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture. EMBO J. 18, 1447–1458 ( 1999).The crystal structure of T4 endonuclease VII, showing an unusual architecture.
Pöhler, J. R. G., Giraud-Panis, M.-J. E. & Lilley, D. M. J. T4 endonuclease VII selects and alters the structure of the four-way DNA junction; binding of a resolution-defective mutant enzyme . J. Mol. Biol. 260, 678– 696 (1996).
Giraud-Panis, M.-J. E., Duckett, D. R. & Lilley, D. M. J. The modular character of a DNA junction resolving enzyme: a zinc binding motif in T4 endonuclease VII. J. Mol. Biol. 252, 596–610 ( 1995).
Birkenbihl, R. P. & Kemper, B. Localization and characterization of the dimerization domain of Holliday structure resolving endonuclease VII of phage T4. J. Mol. Biol. 280, 73–83 (1998).
Giraud-Panis, M.-J. E. & Lilley, D. M. J. T4 endonuclease VII: importance of a histidine-aspartate cluster within the zinc-binding domain . J. Biol. Chem. 271, 33148– 33155 (1996).
Golz, S., Christoph, A., Birkenkamp Demtroder, K. & Kemper, B. Identification of amino acids of endonuclease VII essential for binding and cleavage of cruciform DNA. Eur. J. Biochem. 245, 573–580 (1997).
Hadden, J. M., Convery, M. A., Déclais, A.-C., Lilley, D. M. J. & Phillips, S. E. V. Crystal structure of the Holliday junction-resolving enzyme T7 endonuclease I at 2.1 Å resolution. Nature Struct. Biol. 8, 62–67 (2001).The crystal structure of T7 endonuclease I, revealing the mixed active site.
Guo, F., Gopaul, D. N. & Van Duyne, G. D. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389, 40–46 (1997).
Déclais, A.-C., Hadden, J. M., Phillips, S. E. V. & Lilley, D. M. J. The active site of the junction-resolving enzyme T7 endonuclease I. J. Mol. Biol. 307, 1145–1158 (2001).
Chen, Y., Narendra, U., Iype, L. E., Cox, M. M. & Rice, P. A. Crystal structure of a Flp recombinase–Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell 6, 885–897 ( 2000).
Bond, C. S., Kvaratskhelia, M., Richard, D., White, M. F. & Hunter, W. N. Structure of Hjc, a Holliday junction resolvase, from Sulfolobus solfataricus. Proc. Natl Acad. Sci. USA 98, 5509–5514 ( 2001).The crystal structure of the archaeal Hjc enzyme confirms its close relationship with the type II restriction enzymes and allows prediction of a model for junction binding and cleavage.
Nishino, T., Komori, K., Tsuchiya, D., Ishino, Y. & Morikawa, K. Crystal structure of the archaeal Holliday junction resolvase Hjc and implications for DNA recognition. Structure 9, 197–204 (2001).
White, M. F. & Lilley, D. M. J. Characterization of a Holliday junction resolving enzyme from Schizosaccharomyces pombe. Mol. Cell. Biol. 17, 6465–6471 (1997).
Venclovas, C. & Siksnys, V. Different enzymes with similar structures involved in Mg2+-mediated polynucleotidyl transfer. Nature Struct. Biol. 2, 838–841 (1995).
Ristriani, T. et al. HPV oncoprotein E6 is a structure-dependent DNA-binding protein that recognizes four-way junctions. J. Mol. Biol. 296 , 1189–1203 (2000).
Sharples, G. J., Ingleston, S. M. & Lloyd, R. G. Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J. Bacteriol. 181, 5543–5550 (1999).
Parkinson, M. J. & Lilley, D. M. J. The junction-resolving enzyme T7 endonuclease I: quaternary structure and interaction with DNA. J. Mol. Biol. 270, 169–178 (1997).
White, M. F. & Lilley, D. M. J. The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J. Mol. Biol. 257, 330– 341 (1996).
Giraud-Panis, M.-J. E. & Lilley, D. M. J. Structural recognition and distortion by the DNA junction-resolving enzyme RusA. J. Mol. Biol. 278, 117–133 (1998).
White, M. F. & Lilley, D. M. J. The resolving enzyme CCE1 of yeast opens the structure of the four-way DNA junction. J. Mol. Biol. 266, 122–134 ( 1997).
Kosak, H. G. & Kemper, B. W. Large-scale preparation of T4 endonuclease VII from over-expressing bacteria. Eur. J. Biochem. 194, 779–784 ( 1990).
Iwasaki, H., Takahagi, M., Shiba, T., Nakata, A. & Shinagawa, H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 10, 4381–4389 (1991).
Duckett, D. R., Giraud-Panis, M.-E. & Lilley, D. M. J. Binding of the junction-resolving enzyme bacteriophage T7 endonuclease I to DNA: separation of binding and catalysis by mutation . J. Mol. Biol. 246, 95– 107 (1995).
Bennett, R. J. & West, S. C. Structural analysis of the RuvC–Holliday junction complex reveals an unfolded junction. J. Mol. Biol. 252, 213–226 (1995).
White, M. F. & Lilley, D. M. J. Interaction of the resolving enzyme YDC2 with the four-way DNA junction. Nucleic Acids Res. 26, 5609–5616 ( 1998).
Déclais, A.-C. & Lilley, D. M. J. Extensive central disruption of a four-way junction on binding CCE1 resolving enzyme . J. Mol. Biol. 296, 421– 433 (2000).Cce1 breaks the central base pairs of the junction upon binding, shown by large increases in the fluorescent quantum yield of 2-aminopurine selectively incorporated at the centre.
Shah, R., Bennett, R. J. & West, S. C. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell 79, 853–864 ( 1994).
Schofield, M. J., Lilley, D. M. J. & White, M. F. Dissection of the sequence specificity of the Holliday junction endonuclease CCE1. Biochemistry 37, 7733–7740 (1998).
Fogg, J. M., Schofield, M. J., White, M. F. & Lilley, D. M. J. Sequence and functional-group specificity for cleavage of DNA junctions by RuvC of Escherichia coli. Biochemistry 38, 11349–11358 (1999).
Parkinson, M. J., Pöhler, J. R. G. & Lilley, D. M. J. Catalytic and binding mutants of the junction-resolving enzyme endonuclease I of bacteriophage T7: role of acidic residues. Nucleic Acids Res. 27, 682–689 (1999).
Bolt, E. L., Sharples, G. J. & Lloyd, R. G. Identification of three aspartic acid residues essential for catalysis by the RusA Holliday junction resolvase. J. Mol. Biol. 286, 403–415 ( 1999).
Wardleworth, B. N., Kvaratskhelia, M. & White, M. F. Site-directed mutagenesis of the yeast resolving enzyme Cce1 reveals catalytic residues and relationship with the intron-splicing factor Mrs1. J. Biol. Chem. 275, 23725– 23728 (2000).
Komori, K. et al. Mutational analysis of the Pyrococcus furiosus Holliday junction resolvase hjc revealed functionally important residues for dimer formation, junction DNA binding, and cleavage activities. J. Biol. Chem. 275, 40385–40391 ( 2000).
Kvaratskhelia, M., George, S. J., Cooper, A. & White, M. F. Quantitation of metal ion and DNA junction binding to the Holliday junction endonuclease Cce1. Biochemistry 38, 16613 –16619 (1999).
Daiyasu, H., Komori, K., Sakae, S., Ishino, Y. & Toh, H. Hjc resolvase is a distantly related member of the type II restriction endonuclease family. Nucleic Acids Res. 28, 4540– 4543 (2000).
Makarova, K. S., Aravind, L. & Koonin, E. V. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories . Nucleic Acids Res. 28, 3417– 3432 (2000).
Hickman, A. B. et al. Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell 5, 1025–1034 (2000).
Wah, D. A., Hirsch, J. A., Dorner, L. F., Schildkraut, I. & Aggarwal, A. K. Structure of the multimodular endonuclease FokI bound to DNA. Nature 388, 97–100 (1997).
Giraud-Panis, M.-J. E. & Lilley, D. M. J. Near-simultaneous DNA cleavage by the subunits of the junction-resolving enzyme T4 endonuclease VII. EMBO J. 16, 2528–2534 (1997).
Fogg, J. M., Schofield, M. J., Déclais, A.-C. & Lilley, D. M. J. The yeast resolving enzyme CCE1 makes sequential cleavages in DNA junctions within the lifetime of the complex. Biochemistry 39 , 4082–4089 (2000).
Birkenbihl, R. P. & Kemper, B. Endonuclease VII has two DNA-binding sites each composed from one N- and one C-terminus provided by different subunits of the protein dimer. EMBO J. 17, 4527–4534 (1998).
Shah, R., Cosstick, R. & West, S. C. The RuvC protein dimer resolves Holliday junctions by a dual incision mechanism that involves base-specific contacts. EMBO J. 16, 1464–1472 ( 1997).
Fogg, J. M. & Lilley, D. M. J. Ensuring productive resolution by the junction-resolving enzyme RuvC: Large enhancement of second-strand cleavage rate. Biochemistry 39, 16125– 16134 (2001).
Lilley, D. M. J. et al. Nomenclature Committee of the International Union of Biochemistry: a nomenclature of junctions and branchpoints in nucleic acids. Recommendations 1994. Eur. J. Biochem. 230, 1–2 (1995).
Clegg, R. M., Murchie, A. I. H., Zechel, A. & Lilley, D. M. J. The solution structure of the four-way DNA junction at low salt concentration; a fluorescence resonance energy transfer analysis. Biophys. J. 66, 99–109 ( 1994).
Churchill, M. E., Tullius, T. D., Kallenbach, N. R. & Seeman, N. C. A Holliday recombination intermediate is twofold symmetric. Proc. Natl Acad. Sci. USA 85, 4653–4656 (1988).
Cooper, J. P. & Hagerman, P. J. Geometry of a branched DNA structure in solution. Proc. Natl Acad. Sci. USA 86, 7336–7340 (1989).
Murchie, A. I. H. et al. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature 341, 763–766 ( 1989).
Nowakowski, J., Shim, P. J., Prasad, G. S., Stout, C. D. & Joyce, G. F. Crystal structure of an 82 nucleotide RNA–DNA complex formed by the 10–23 DNA enzyme. Nature Struct. Biol. 6, 151–156 (1999).
Ortiz-Lombardía, M. et al. Crystal structure of a DNA Holliday junction. Nature Struct. Biol. 6, 913–917 (1999).
Eichman, B. F., Vargason, J. M., Mooers, B. H. M. & Ho, P. S. The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions. Proc. Natl Acad. Sci. USA 97, 3971–3976 ( 2000).The crystal structure of a four-way DNA junction in the stacked X-shape conformation. This is the first structure that is completely free of base mismatches.
Miick, S. M., Fee, R. S., Millar, D. P. & Chazin, W. J. Crossover isomer bias is the primary sequence-dependent property of immobilized Holliday junctions. Proc. Natl Acad. Sci. USA 94, 9080–9084 (1997).
Grainger, R. J., Murchie, A. I. H. & Lilley, D. M. J. Exchange between stacking conformers in a four-way DNA junction. Biochemistry 37, 23– 32 (1998).
Lilley, D. M. J. Structures of helical junctions in nucleic acids. Quart. Rev. Biophys. 33, 109–159 ( 2000).
Author information
Authors and Affiliations
Related links
Related links
DATABASE LINKS
FURTHER INFORMATION
ENCYCLOPEDIA OF LIFE SCIENCES
Eukaryotic recombination: initiation by double-strand breaks
Glossary
- BRANCH MIGRATION
-
The process of exchange of base-pairing partners at a helical junction formed from homologous sequences.
- INTEGRASE
-
The integrase protein of phage-λ is a tyrosine-recombinase protein that mediates the site-specific integration of the phage DNA into the host genome.
- HETERODUPLEX
-
A DNA duplex formed by association between two homologous strands, each of which was previously hybridized to different complements. If the homology is less than 100%, the heteroduplex will contain base mismatches that will require repair.
- GENETIC DRIFT
-
Random changes occurring in a gene over time, often with little or no phenotype.
- RESTRICTION ENDONUCLEASES
-
Sequence-specific nucleases used by bacteria in defence against phage infection. These can be subclassified into different types according to their target and mode of action.
- SUBUNIT-EXCHANGE RATES
-
In free solution, the subunits of junction-resolving enzymes may exchange between dimers. In general, this is characterized by a single exponential, giving the half-time for the exchange process. These vary widely for the known junction-resolving enzymes.
- pI VALUE
-
The pH at which the ionization state of charged amino-acid side chains results in an overall electrical neutrality.
- TRANSITION STATE
-
The point of highest energy along a reaction potential energy surface.
- ELECTRON PARAMAGNETIC RESONANCE SPECTROSCOPY
-
(EPR spectroscopy). Observation of the transitions between spin states of an unpaired electron in a magnetic field. The unpaired 3d5 electrons of manganese can be observed by EPR, providing information on the chemical environment of the metal ion.
- ISOTHERMAL TITRATION CALORIMETRY
-
(ITC). Involves the measurement of heat absorbed or evolved as two interacting components are titrated in a cell. This provides a direct measure of the reaction enthalpy. The association constant can also be measured, from which other thermodynamic parameters can be calculated.
- TN7 TRANSPOSASE
-
A polynucleotide transferase involved in the transposition of Tn7.
- LEWIS ACID
-
A broader definition of acidity that encompasses any positively charged group, such as a metal cation.
Rights and permissions
About this article
Cite this article
Lilley, D., White, M. The junction-resolving enzymes. Nat Rev Mol Cell Biol 2, 433–443 (2001). https://doi.org/10.1038/35073057x
Issue Date:
DOI: https://doi.org/10.1038/35073057x
This article is cited by
-
Junction resolving enzymes use multivalency to keep the Holliday junction dynamic
Nature Chemical Biology (2019)
-
Structural basis of sequence-specific Holliday junction cleavage by MOC1
Nature Chemical Biology (2019)
-
CRISPR–Cas adaptation: insights into the mechanism of action
Nature Reviews Microbiology (2016)
-
Mutation and association analysis of GEN1 in breast cancer susceptibility
Breast Cancer Research and Treatment (2010)
-
Identification of Holliday junction resolvases from humans and yeast
Nature (2008)