Key Points
-
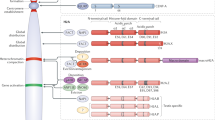
Chromatin is composed of nucleosomes, which consist of DNA and histone proteins. The histone amino terminus has a 'tail' region that is highly conserved between species and consists of protein modules that are responsible for post-translational modification of histones.
-
Many histone modifications take place in the form of acetylation, methylation or phosphorylation, and have been correlated with nuclear activities such as replication, chromatin assembly and transcription. It is thought that modification of histones is a sequential or combinatorial process such that a 'histone code' materializes which promotes transcriptional events downstream.
-
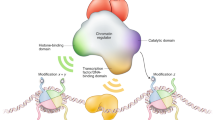
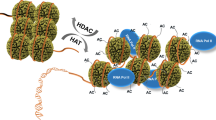
Histone acetylation/deacetylation is the most studied histone modification activity. It is correlated with transcriptional activation/repression. The enzymes that mediate this process are histone acetyltransferases (HATs) and histone deacetylases (HDACs). Bromodomain and kinase domain modules can act as transcriptional coactivators by aiding the process to proceed. There are distinct functional links between HATs/HDACs and these modules.
-
Gene silencing occurs during the formation of higher-order heterochromatin through histone methylation. This process is aided by SET domains and chromodomains.
-
Other conserved protein modules involved in histone modification are also known, although their biochemical functions are as yet poorly defined. Of those modules that have been studied extensively, it is still not entirely clear what the function of these modules is or in what sequence they interact with histone tails to coordinate and regulate gene expression. Futher studies will concentrate on these issues.
Abstract
Histones are the predominant protein components of chromatin and are subject to specific post-translational modifications that are correlated with transcriptional competence. Among these histone modifications are acetylation, phosphorylation and methylation, and recent studies reveal that conserved protein modules mediate the attachment, removal or recognition of these modifications. It is becoming clear that appropriate coordination of histone modifications and their manipulations by conserved protein modules are integral to gene-specific transcriptional regulation within chromatin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wolffe, A. P. Chromatin: Structure and Function (Academic Press, London, 1992).
Allfrey, V. G., Faulkner, R. & Mirsky, A. E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA 51, 786–794 (1964).
Bradbury, E. M. Reversible histone modifications and the chromosome cell cycle. BioEssays 14, 9–16 (1992).
Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 (1997).
Thompson, J. S., Ling, X. & Grunstein, M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369, 245–247 (1994).
Durrin, L., Mann, R., Kayne, P. & Grunstein, M. Yeast histone H4 N–terminal sequence is required for promoter activation in vivo. Cell 65, 1023–1031 (1991).
Wolffe, A. P. & Hayes, J. J. Chromatin disruption and modification. Nucleic Acids Res. 27, 711–720 (1999).
Morales, V. & Richard-Foy, H. Role of histone N-terminal tails and their acetylation in nucleosome dynamics. Mol. Cell. Biol. 20, 7230–7237 (2000).
Hansen, J. C., Tse, C. & Wolffe, A. P. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry 37, 17637–17641 (1998).
Sivolob, A., De Lucia, F., Alilat, M. & Prunell, A. Nucleosome dynamics. VI. Histone tail regulation of tetrasome chiral transition. A relaxation study of tetrasomes on DNA minicircles. J. Mol. Biol. 295, 55–69 (2000).
Wang, X., Moore, S. C., Laszckzak, M. & Ausio, J. Acetylation increases the α-helical content of the histone tails of the nucleosome. J. Biol. Chem. 275, 35013–35020 (2000).
Mutskov, V. et al. Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol. Cell. Biol. 18, 6293–6304 (1998).
Polach, K. J., Lowary, P. T. & Widom, J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J. Mol. Biol. 298, 211–223 (2000).
Turner, B. M. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell. Mol. Life Sci. 54, 21–31 (1998).The first to suggest that histone tail modifications may be histone marks that signal downstream transcriptional events.
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).Builds on the initial proposal of Turner, to propose that distinct histone tail modifications provide a 'histone code' for downstream transcriptional activities.
Georgakopoulos, T. & Thireos, G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 11, 4145–4152 (1992).
Marcus, G., Silverman, N., Berger, S., Horiuchi, J. & Guarente, L. Functional similarity and physical association between GCN5 and ADA2-putative transcriptional adaptors. EMBO J. 13, 4807–4815 (1994).
Horiuchi, J., Silverman, N., Marcus, G. A. & Guarente, L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol. Cell. Biol. 15, 1203–1209 (1995).
Candau, R. & Berger, S. L. Structural and functional analysis of yeast putative adaptors: evidence for an adaptor complex in vivo. J. Biol. Chem. 271, 5237–5345 (1996).
Barlev, N. A. et al. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270, 19337–19344 (1995).
Silverman, N., Agapite, J. & Guarente, L. Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription. Proc. Natl Acad. Sci. USA 91, 11665–11668 (1994).
Zamir, I. et al. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol. Cell. Biol. 16, 5458–5465 (1996).
Horlein, A. J. et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377, 397–404 (1995).
Kurokawa, R. et al. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature 377, 451–454 (1995).
Heinzel, T. et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387, 43–48 (1997).
Alland, L. et al. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387, 49–55 (1997).
Pogo, B. G. T., Allfrey, V. G. & Mirsky, A. E. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc. Natl Acad. Sci. USA 55, 6212–6222 (1966).
Vidali, G., Boffa, L. C., Bradbury, E. M. & Allfrey, V. G. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNAase I sensitivity of the associated DNA sequences. Proc. Natl Acad. Sci. USA 75, 2239–2243 (1978).
Hebbes, T. R., Thorne, A. W. & Crane-Robinson, C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7, 1395–1403 (1988).
Hebbes, T. R., Clayton A. L., Throne A. W. & Crane-Robinson, C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 13, 1823–1830 (1994).
Vettese-Dadey, M. et al. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J 15, 2508–2518 (1996).
Sealy, L. & Chalkley, R. DNA associated with hyperacetylated histone is preferentially digested by DNase I. Nucleic Acids Res. 5, 1863–1876 (1978).
Brownell, J. E. et al. Tetrahymena histone acetyltransferase A: a homolog of yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851 (1996).The first to functionally link gene activation by a transcriptional co-activator to the histone acetyltransferase activity of that co-activator.
Berger, S. L. et al. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70, 251–265 (1992).
Wang, L., Liu, L. & Berger, S. L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12, 640–653 (1998).
Kuo, M. H., Zhou, J. X., Jambeck, P., Churchill, M. E. A. & Allis, C. D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12, 627–639 (1998).
Shikama, N., Lyon, J. & LaThangue, N. B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7, 230–236 (1997).
Bannister, A. J. & Kouzarides, T. The CBP co-activator is a histone acetyltransferase. Nature 384, 641–643 (1996).
Mizzen, C. A. et al. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell 87, 1261–1270 (1996).
Cress, W. D. & Seto, E. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184, 1–16 (2000).
Kuo, M. H. & Allis, C. D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20, 615–626 (1998).
Neal, K. C., Pannuti, A., Smith, E. R. & Lucchesi, J. C. A new human member of the MYST family of histone acetyl transferases with high sequence similarity to Drosophila MOF. Biochim. Biophys. Acta1490, 170–174 (2000).
Sterner, D. E. & Berger, S. L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459 (2000).
Kawasaki, H. et al. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature 405, 195–200 (2000).
Takechi, S. & Nakayama, T. Sas3 is a histone acetyltransferase and requires a zinc finger motif. Biochem. Biophys. Res. Commun. 266, 405–410 (1999).
EhrenhoferMurray, A. E., Rivier, D. H. & Rine, J. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics 145, 923–934 (1997).
Kuo, M. H. et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383, 269–272 (1996).
Trievel, R. C., Li, F.-Y. & Marmorstein, R. Application of a novel fluorescence histone acetyltransferase enzyme assay to study the substrate specificity of human PCAF. Anal. Biochem. 287, 319–328 (2000).
John, S. et al. The Something About Silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAFII30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)–FACT complex. Genes Dev. 14, 1196–1208 (2000).
Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 (1996).
Grant, P. A. et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640–1650 (1997).
Allard, S. et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18, 5108–5119 (1999).
Grant, P. A. et al. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895–5900 (1999).
Trievel, R. C. et al. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl Acad. Sci. USA 96, 8931–8936 (1999).
Clements, A. et al. Crystal structure of the histone acetyltransferase domain of the human P/CAF transcriptional regulator bound to coenzyme-A. EMBO J. 18, 3521–3532 (1999).
Rojas, J. R. et al. Structure of the Tetrahymena GCN5 bound to coenzyme-A and a histone H3 peptide. Nature 401, 93–98 (1999).Revealed the first high-resolution structure of a HAT bound to a histone peptide substrate.
Lin, Y., Fletcher, C. M., Zhou, J., Allis, C. D. & Wagner, G. Solution structure of the catalytic domain of Tetrahymena GCN5 histone acetyltransferase in complex with coenzyme A. Nature 400, 86–89 (1999).
Yan, Y., Barlev, N. A., Haley, R. H., Berger, S. L. & Marmorstein, R. Crystal structure of yeast Esa1 suggests a unified mechanism of catalysis and substrate binding by histone acetyltransferases. Mol. Cell 6, 1195–1205 (2000).
Dutnall, R. N., Tafrov, S. T., Sternglanz, R. & Ramakrishnan, V. Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell 94, 427–438 (1998).
Jeanmougin, F., Wurtz, J. M., LeDouarin, B., Chambon, P. & Losson, R. The bromodomain revisited. Trends Biochem. Sci. 22, 151–153 (1997).
Winston, F. & Allis, C. D. The bromodomain: a chromatin-targeting module? Nature Struct. Biol. 6, 601–604 (1999).
Sterner, D. E. et al. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19, 86–98 (1999).
Kraus, W. L., Manning, E. T. & Kadonaga, J. T. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19, 8123–8135 (1999).
Du, J., Nasir, I., Benton, B. K., Kladde, M. P. & Laurent, B. C. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics 150, 987–1005 (1998).
Matangkasombut, O., Buratowski, R. M., Swilling, N. W. & Buratowski, S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev 14, 951–962 (2000).
Ornaghi, P., Ballario, P., Lena, A. M., Gonzalez, A. & Filetici, P. The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J. Mol. Biol. 287, 1–7 (1999).
Dhalluin, C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496 (1999).Provided the first evidence that bromodomains specifically target acetyl-lysine-containing histone substrates.
Jacobson, R. H., Ladurner, A. G., King, D. S. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000).
Owen, D. J. et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 19, 6141–6149 (2000).
Clayton, A. L., Rose, S., Barratt, M. J. & Mahadevan, L. C. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 19, 3714–3726 (2000).
Nowak, S. J. & Corces, V. G. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 14, 3003–3013 (2000).
Cheung, P. et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5, 905–915 (2000).
Lo, W. -S. et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5, 917–926 (2000).References 72 and 73 revealed that Ser10 phosphorylation of histone H3 is functionally linked to histone acetylation at Lys14 and to gene activation of a subset of Gcn5-dependent genes.
Hsu, J. Y. et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/Aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 (2000).
De Souza, C. P., Osmani, A. H., Wu, L. P., Spotts, J. L. & Osmani, S. A. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102, 293–302 (2000).References 74 and 75 indicate that phosphorylation of histone H3 at Ser 10 is mediated by the Ipl1/Aurora kinase and that this activity is functionally linked to chromosome condensation during mitosis.
Hassan, A. H., Neely, K. E. & Workman, J. L. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104, 817–827 (2001).
Cosma, M. P., Tanaka, T. & Nasmyth, K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97, 299–311 (1999).
Krebs, J. E., Fry, C. J., Samuels, M. L. & Peterson, C. L. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102, 587–598 (2000).References 76 – 78 indicate a functional link between histone acetylation and chromatin remodelling.
Weiler, K. S. & Wakimoto, B. T. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29, 577–605 (1995).
Grunstein, M. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 9, 383–387 (1997).
Reuter, G. & Spierer, P. Position effect variegation and chromatin proteins. Bioessays 14, 605–612 (1992).
Allshire, R. C., Nimmo, E. R., Ekwall, K., Javerzat, J. P. & Cranston, G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9, 218–233 (1995).
Schotta, G. & Reuter, G. Controlled expression of tagged proteins in Drosophila using a new modular P-element vector system. Mol. Gen. Genet. 262, 916–920 (2000).
Tschiersch, B. et al. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13, 3822–3831 (1994).
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).Shows that suppressors of variegation gene product, Su(var)3–9, mediates gene silencing through its histone H3 methyltransferase activity.
Jenuwein, T., Laible, G., Dorn, R. & Reuter, G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci. 54, 80–93 (1998).
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 401, 120–124 (2001).
Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 401, 116–120 (2001).References 87 and 88 indicate that the chromodomain of the heterochromatin-associated protein, HP1, targets the Lys9 of histone H3 for heterochromatin assembly and gene silencing.
Eissenberg, J. C. et al. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 87, 9923–9927 (1990).
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001).
Akhtar, A., Zink, D. & Becker, P. B. Chromodomains are protein–RNA interaction modules. Nature 407, 405–409 (2000).
Ball, L. J. et al. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 16, 2473–2481 (1997).Provides the first atomic structure of a chromodomain.
Platero, J. S., Hartnett, T. & Eissenberg, J. C. Functional analysis of the chromo domain of HP1. EMBO J. 14, 3977–3986 (1995).
Edmondson, S. P., Qiu, L. & Shriver, J. W. Solution structure of the DNA-binding protein Sac7d from the hyperthermophile Sulfolobus acidocaldarius. Biochemistry 34, 13289–133304 (1995).
Baumann, H., Knapp, S., Lundback, T., Ladenstein, R. & Hard, T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nature Struct. Biol. 1, 808–819 (1994).
Aasland, R. & Stewart, A. F. The chrome shadow domain, a second chrome domain in heterochromatin-binding protein-1, HP1. Nucleic Acids Res. 23, 3168–3173 (1995).
Fanti, L., Giovinazzo, G., Berloco, M. & Pimpinelli, S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2, 527–538 (1998).
Murzina, N., Verreault, A., Laue, E. & Stillman, B. Heterochomatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4, 529–540 (1999).
Smothers, J. F. & Henikoff, S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 10, 27–30 (2000).
Brasher, S. V. et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chrome domain dimer. EMBO J. 19, 1587–1597 (2000).Provides the first atomic structure of a chromoshadow domain.
Strahl, B. D., Ohba, R., Cook, R. G. & Allis, C. D. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl Acad. Sci. USA 96, 14967–14972 (1999).Indicates that histone methylation can be correlated with gene activation.
Chen, D. et al. Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177 (1999).
Stallcup, M. R. et al. Co-operation between protein-acetylating and protein-methylating co-activators in transcriptional activation. Biochem. Soc. Trans. 28, 415–418 (2000).
Travers, A. An engine for nucleosome remodeling. Cell 96, 311–314 (1999).
Carlson, M. & Laurent, B. C. The SNF/SWI family of global transcriptional activators. Curr. Opin. Cell Biol. 6, 396–402 (1994).
Peterson, C. L. & Tamkun, J. W. The SWI–SNF complex: a chromatin remodeling machine? Trends Biochem. Sci. 20, 143–146 (1995).
Ng, H. H. & Bird, A. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25, 121–126 (2000).
Kouzarides, T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet. Dev. 9, 40–48 (1999).
Xue, Y. et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861 (1998).Describes a transcriptional regulatory complex that has both histone deacetylase and chromatin remodelling activity, indicating a direct link between the histone acetylation status and chromatin remodelling.
Borden, K. L. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295, 1103–1112 (2000).
Aasland, R., Gibson, T. J. & Stewart, A. F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20, 56–59 (1995).
Jackson, P. K. et al. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10, 429–439 (2000).
Aasland, R., Stewart, A. F. & Gibson, T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional corepressor N-CoR and TFIIIB. Trends Biochem. Sci. 21, 87–88 (1996).
Sullivan, S. A., Aravind, L., Makalowski, I., Baxevanis, A. D. & Landsman, D. The histone database: a comprehensive WWW resource for histones and histone fold-containing proteins. Nucleic Acids Res. 28, 320–322 (2000).
Burley, S. K. & Roeder, R. G. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65, 769–799 (1996).
Xie, X. et al. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380, 316–322 (1996).
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 389, 251–260 (1997).Describes the atomic structure of the nucleosome core particle indicating that, at least in the context of the core particle, the histone tail regions are largely disordered.
Author information
Authors and Affiliations
Related links
Related links
DATABASE LINKS
Glossary
- NUCLEAR HORMONE RECEPTOR COREPRESSORS
-
Proteins that interact with DNA-bound nuclear hormone receptors to facilitate the repression of gene expression.
- HETEROCHROMATIN
-
A highly condensed form of chromatin that occurs at defined sites, such as silencer DNA elements or telomeres.
- BROMODOMAIN
-
A domain with sequence conservation that is found in several transcriptional regulatory proteins involved in gene activation, and contains acetyl-lysine binding activity.
- IPL1/AURORA KINASES
-
A family of homologous kinases in which histone H3 kinase activity is correlated with chromosome condensation.
- TELOMERES
-
The ends of eukaryotic chromosomes.
- EUCHROMATIC
-
DNA that contains most of the structural genes, changes structure during the cell cycle and undergoes transcriptional regulation.
- MATING TYPE LOCI
-
Gene regulatory regions of the yeast chromosome that control the sexual fate of the yeast cell.
- CHROMODOMAIN
-
A domain with sequence conservation that is found in several transcriptional regulatory proteins. The chromodomain from HP1 has been shown to contain methyl-lysine-binding activity.
- DOSAGE COMPENSATION
-
A process in organisms that have male and female sexes, which ensures that different copies of the sex chromosome between these sexes have the same amount of most of the sex chromosome gene products.
- HISTONE FOLD
-
Refers to proteins that contain the same tertiary structure as the core region of the core histones.
- CHROMOSHADOW DOMAIN
-
A domain in HP1 that is highly related in sequence and structure to the chromodomain. Unlike the chromodomain, which is monomeric, the chromoshadow domain is dimeric.
Rights and permissions
About this article
Cite this article
Marmorstein, R. Protein modules that manipulate histone tails for chromatin regulation. Nat Rev Mol Cell Biol 2, 422–432 (2001). https://doi.org/10.1038/35073047
Issue Date:
DOI: https://doi.org/10.1038/35073047
This article is cited by
-
Targeting PDZ domains as potential treatment for viral infections, neurodegeneration and cancer
Biology Direct (2021)
-
Airway hyperresponsiveness development and the toxicity of PM2.5
Environmental Science and Pollution Research (2021)
-
Tri-methylation of ATF7IP by G9a/GLP recruits the chromodomain protein MPP8
Epigenetics & Chromatin (2018)
-
Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo
Oncogenesis (2015)
-
Epigenetic regulation of mucin genes in human cancers
Clinical Epigenetics (2011)