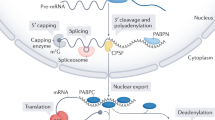
Key Points
-
Messenger RNA abundance is determined by balancing transcription and RNA decay. mRNA stability can be rapidly modulated to alter the expression of specific genes thereby providing flexibility in effecting changes in patterns of protein synthesis.
-
An evolutionarily conserved mRNA-degradation pathway is initiated by the removal of the 3′ poly(A) tail. This disrupts the translation initiation complex and provides degradative enzymes with access to the 5′ cap and remaining RNA body.
-
The intimate relationship between mRNA decay and translation is further indicated by the ability of translation-initiation factors and proteins that bind the poly(A) tail to protect the mRNA from degradation. Moreover, evidence shows that inhibiting translation elongation promotes mRNA stabilization
-
The turnover of mRNAs is also regulated by cis-acting elements that either promote or inhibit their decay. The most prevalent is the A+U-rich element (ARE), found in the 3′untranslated region (3′ UTR) of mRNAs encoding many important growth control proteins. ARE-binding proteins affect mRNA stability, translation and subcellular localization. Other elements found in the 5′ UTR and coding regions also modulate transcript stability.
-
Several signalling pathways are implicated in triggering changes in stability of specific mRNAs. One example is the interleukin-2 mRNA, which is stabilized by the c-Jun amino-terminal kinase (JNK) signalling pathway through JNK-responsive elements in its 5′UTR.
-
A strong link between translation and RNA turnover is also shown by nonsense-mediated decay, which ensures that mRNAs containing premature stop codons are degraded.
Abstract
The levels of cellular messenger RNA transcripts can be regulated by controlling the rate at which the mRNA decays. Because decay rates affect the expression of specific genes, they provide a cell with flexibility in effecting rapid change. Moreover, many clinically relevant mRNAs — including several encoding cytokines, growth factors and proto-oncogenes — are regulated by differential RNA stability. But what are the sequence elements and factors that control the half-lives of mRNAs?
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Herrick, D., Parker, R. & Jacobson, A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 2269–2284 (1990).
Proudfoot, N. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 25, 290–293 ( 2000).
Chen, C. Y. & Shyu, A. B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465–470 (1995).
Hilleren, P. & Parker, R. Mechanisms of mRNA surveillance in eukaryotes. Annu. Rev. Genet. 33, 229– 260 (1999).
He, W. & Parker, R. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 12 , 346–350 (2000).
Conne, B., Stutz, A. & Vassalli, J. D. The 3′ untranslated region of messenger RNA: a molecular 'hotspot' for pathology? Nature Med. 6, 637–641 (2000).
Brown, C. E. & Sachs, A. B. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 18, 6548–6559 (1998).
Boeck, R. et al. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 271, 432–438 (1996).
Tucker, M. et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae . Cell 104, 377–386 (2001).
Chang, M. et al. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19, 1056–1067 ( 1999).
LaGrandeur,T. E. & Parker, R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17, 1487–1496 (1998).
Beelman, C. A. et al. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382, 642–646 (1996).
Dunckley, T. & Parker, R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18, 5411– 5422 (1999).
Zhang, S., Williams, C. J., Hagan, K. & Peltz, S. W. Mutations in VPS16 and MRT1 stabilize mRNAs by activating an inhibitor of the decapping enzyme. Mol. Cell. Biol. 19, 7568–7576 (1999).
Hatfield, L., Beelman, C. A., Stevens, A. & Parker, R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 5830 –5838 (1996).
Bonnerot, C., Boeck, R. & Lapeyre, B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p . Mol. Cell. Biol. 20, 5939– 5946 (2000).
Tharun, S. et al. Yeast Sm-like proteins function in mRNA decapping and decay . Nature 404, 515–518 (2000).
Bouveret, E., Rigaut, G., Shevchenko, A., Wilm, M. & Seraphin, B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19, 1661–1671 (2000).
Hsu, C. L. & Stevens, A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13 , 4826–4835 (1993).
Jacobs, J. S., Anderson, A. R. & Parker, R. P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17, 1497–1506 ( 1998).
Shyu, A. B., Belasco, J. G. & Greenberg, M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5, 221–231 ( 1991).
Korner, C. G. et al. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 17, 5427–5437 ( 1998).
Korner, C. G. & Wahle, E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J. Biol. Chem. 272, 10448–10456 (1997).
Martinez, J. et al. A 54-kDa fragment of the poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting poly(A)-specific 3′ exonuclease . J. Biol. Chem. 275, 24222– 24230 (2000).
Couttet, P., Fromont-Racine, M., Steel, D., Pictet, R. & Grange, T. Messenger RNA deadenylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA 94, 5628–5633 (1997).
Gao, M., Wilusz, C. J., Peltz, S. W. & Wilusz, J. A novel mRNA decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20, 1134– 1143 (2001).The first biochemical evidence for an mRNA-decapping activity in higher eukaryotes. Also the first demonstration that A+U-rich elements can promote decapping as well as deadenylation.
Achsel, T. et al. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18, 5789– 5802 (1999).
Bashkirov, V. I., Scherthan, H., Solinger, J. A., Buerstedde, J. M. & Heyer, W. D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136, 761–773 (1997).
Caruccio, N. & Ross, J. Purification of a human polyribosome-associated 3′ to 5′ exoribonuclease. J. Biol. Chem. 269, 31814–31821 (1994).
Kwan, C. N. A cytoplasmic exoribonuclease from HeLa cells. Biochim. Biophys. Acta 479, 322–331 ( 1977).
Brouwer, R. et al. Three novel components of the human exosome. J. Biol. Chem. 276, 6177–6184 (2001).
Bernstein, P., Peltz, S. W. & Ross, J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 9, 659–670 ( 1989).
Gingras, A. C., Raught, B. & Sonenberg, N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963 ( 1999).
Wells, S. E., Hillner, P. E., Vale, R. D. & Sachs, A. B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140 ( 1998).
Gao, M., Fritz, D. T., Ford, L. P. & Wilusz, J. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell 5, 479–488 (2000). Shows that the mammalian deadenylase is a cap-binding protein and that its activity is stimulated by interaction with the cap in vitro (also see reference 36).
Dehlin, E., Wormington, M., Korner, C. G. & Wahle, E. Cap-dependent deadenylation of mRNA. EMBO J. 19, 1079–1086 (2000). Shows that mammalian and Xenopus PARN is a cap-binding protein and that activity is stimulated by interaction with the cap in vivo and in vitro (also see reference 35).
Ford, L. P., Watson, J., Keene, J. D. & Wilusz, J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 13, 188– 201 (1999).
Caponigro, G. & Parker, R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 9, 2421–2432 ( 1995).
Wormington, M., Searfoss, A. M. & Hurney, C. A. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 15, 900– 909 (1996).
Schwartz, D. C. & Parker, R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 5247–5256 (1999).
Beelman, C. A. & Parker, R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 269, 9687–9692 (1994)
Boeck, R., Lapeyre, B., Brown, C. E. & Sachs, A. B. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 18, 5062– 5072 (1998).
Schwartz, D. C. & Parker, R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 20, 7933–7942 (2000).
Vilela, C., Velasco, C., Ptushkina, M. & McCarthy, J. E. The eukaryotic mRNA decapping protein dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J. 19, 4372–4382 (2000).
Bakheet, T., Frevel, M., Williams, B. R., Greer, W. & Khabar, K. S. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29, 246– 254 (2001).A comprehensive database of A+U-rich elements found in 3′ UTRs of human mRNAs. The database includes both predicted and known AREs and should be an invaluable resource to researchers.
Zhang, W. et al. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13, 7652–7665 (1993).
Myer, V. E., Fan, X. C. & Steitz, J. A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 16, 2130 –2139 (1997).
Piecyk, M. et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19, 4154–4163 (2000).
Lai, W. S. et al. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor-α mRNA. Mol. Cell. Biol. 19, 4311– 4323 (1999).
Peng, S. S., Chen, C. Y., Xu, N. & Shyu, A. B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17, 3461–3470 ( 1998).
DeMaria, C. T. & Brewer, G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem. 271, 12179–12184 (1996).
Loflin, P., Chen, C. Y. & Shyu, A. B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13, 1884–1897 (1999).
Laroia, G., Cuesta, R., Brewer, G. & Schneider, R. J. Control of mRNA decay by heat-shock-ubiquitin–proteasome pathway. Science 284, 499–502 ( 1999).
Carballo, E., Lai, W. S. & Blackshear, P. J. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95, 1891 –1899 (2000).
Taylor, G. A. et al. A pathogenetic role for TNF-α in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency . Immunity 4, 445–454 (1996).
Carballo, E., Lai, W. S. & Blackshear, P. J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281, 1001–1005 (1998).
Wang, X., Kiledjian, M., Weiss, I. M. & Liebhaber, S. A. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability. Mol. Cell. Biol. 15, 1769–1777 (1995).
Wang, Z. & Kiledjian, M. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol. Cell. Biol. 20, 6334–6341 (2000).
Wang, Z., Day, N., Trifillis, P. & Kiledjian, M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro . Mol. Cell. Biol. 19, 4552– 4560 (1999).
Veyrune, J. L., Carillo, S., Vie, A. & Blanchard, J. M. c-fos mRNA instability determinants present within both the coding and the 3′ non coding region link the degradation of this mRNA to its translation . Oncogene 11, 2127–2134 (1995).
Grosset, C. et al. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103, 29–40 (2000).Identification of a multi-protein complex that is required for translation-dependent destabilization of mRNAs containing the c-fos coding region determinant. PABP, PAIP1, Unr, AUF1 and NSAP1 were involved and overexpression of these proteins inhibited deadenylation of mCRD-containing mRNAs.
Chen, C. Y., Gatto-Konczak, F., Wu, Z. & Karin, M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway . Science 280, 1945–1949 (1998).
Chen, C. Y. et al. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 14, 1236–1248 (2000).
Winzen, R. et al. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18, 4969– 4980 (1999).A study of the kinase pathways that are required for ARE-mediated stabilization. The authors use dominant kinase mutants to distinguish between signalling pathways required for mRNA stabilization in vivo.
Lasa, M., Brook, M., Saklatvala, J. & Clark, A. R. Dexamethasone destabilizes cyclooxygenase 2 mRNA by inhibiting mitogen-activated protein kinase p38. Mol. Cell. Biol. 21, 771–780 (2001).
Klein, N., Curatola, A. M. & Schneider, R. J. Calcium-induced stabilization of AU-rich short-lived mRNAs is a common default response. Gene Expr. 7, 357–365 (1999).
Ruiz-Echevarria, M. J. & Peltz, S. W. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101, 741– 751 (2000).
Muhlrad, D. & Parker, R. Premature translational termination triggers mRNA decapping. Nature 370, 578 –581 (1994).
Weng, Y., Czaplinski, K. & Peltz, S. W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 16, 5491–5506 (1996).
Wang, W., Czaplinski, K., Rao, Y. & Peltz, S. W. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20, 880– 890 (2001).
Gonzalez, C. I., Ruiz-Echevarria, M. J., Vasudevan, S., Henry, M. F. & Peltz, S. W. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell 5, 489–499 ( 2000).
Minvielle-Sebastia, L. et al. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 17, 7454–7468 (1998).
Pulak, R. & Anderson, P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7, 1885 –1897 (1993).
Page, M. F., Carr, B., Anders, K. R., Grimson, A. & Anderson, P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast . Mol. Cell. Biol. 19, 5943– 5951 (1999).
Cali, B. M., Kuchma, S. L., Latham, J. & Anderson, P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 151, 605–616 ( 1999).
Serin, G., Gersappe, A., Black, J. D., Aronoff, R. & Maquat, L. E. Identification and characterization of human orthologues to Saccharomyces cerevisiae upf2 protein and upf3 protein (Caenorhabditis elegans SMG-4) Mol. Cell. Biol. 21, 209–223 ( 2001).
Bhattacharya, A. et al. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6, 1226–1235 ( 2000).
Pal, M., Ishigaki, Y., Nagy, E. & Maquat, L. E. Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signalling pathway. RNA 7, 5– 15 (2001).
Sun, X., Perlick, H. A., Dietz, H. C. & Maquat, L. E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA 95, 10009– 10014 (1998).
Mendell, J. T., Medghalchi, S. M., Lake, R. G., Noensie, E. N. & Dietz, H. C. Novel upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 20, 8944– 8957 (2000).Discovery of human and S. pombe homologues of Upf2 allowed the authors to identify regions of the protein with homology to eIF4G. They were able to show interaction of UPF2 with both human eIF4A and human SUI1 by yeast two-hybrid assays.
Lykke-Andersen, J., Shu, M. D. & Steitz, J. A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121–1131 (2000). An elegant demonstration of the role of human Upf proteins in NMD. The authors show that when tethered downstream of a stop codon, Upf proteins can promote NMD of a wild-type transcript and also show that UPF3 is a shuttling protein that associates specifically with spliced mRNAs.
Zhang, J., Sun, X., Qian, Y. & Maquat, L. E. Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4, 801–815 ( 1998).
Kataoka, N. et al. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6, 673–682 (2000).
Le Hir, H., Izaurralde, E., Maquat, L. E. & Moore, M. J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 19, 6860–6869 (2000). Identification of several factors that are deposited on mRNAs by the splicing machinery at a consistent distance from the exon–exon junction. Such proteins are candidates for NMD 'markers'.
Zhou, Z. et al. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407, 401– 405 (2000).
Le Hir, H., Moore, M. J. & Maquat, L. E. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev. 14, 1098–1108 (2000).
McGarvey, T. et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J. Cell Biol. 150, 309–320 (2000).
Fortes, P. et al. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6, 191–196 (2000).
Wyers, F., Minet, M., Dufour, M. E., Vo, L. T. & Lacroute, F. Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol. Cell. Biol. 20, 3538–3549 (2000).
Horazdovsky, B. F. & Emr, S. D. The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J. Biol. Chem. 268 , 4953–4962 (1993).
Wang, X., Watt, P. M., Louis, E. J., Borts, R. H. & Hickson, I. D. Pat1: a topoisomerase II-associated protein required for faithful chromosome transmission in Saccharomyces cerevisiae. Nucleic Acids Res. 24, 4791 –4797 (1996).
Salgado-Garrido, J., Bragado-Nilsson, E., Kandels-Lewis, S. & Seraphin, B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 18, 3451– 3462 (1999).
Dunckley, T., Tucker, M. & Parker, R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157, 27–37 (2001).
Czaplinski, K., Weng, Y., Hagan, K. W. & Peltz, S. W. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1, 610– 623 (1995).
Leeds, P., Peltz, S. W., Jacobson, A. & Culbertson, M. R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 5, 2303–2314 ( 1991).
Applequist, S. E., Selg, M., Raman, C. & Jack, H. M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 25, 814–821 (1997).
He, F., Brown, A. H. & Jacobson, A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17, 1580–1594 ( 1997).
Cui, Y., Hagan, K. W., Zhang, S. & Peltz, S. W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 9, 423–436 ( 1995).
Ponting, C. P. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem. Sci. 25, 423 –426 (2000).
Shirley, R. L., Lelivelt, M. J., Schenkman, L. R., Dahlseid, J. N. & Culbertson, M. R. A factor required for nonsense-mediated mRNA decay in yeast is exported from the nucleus to the cytoplasm by a nuclear export signal sequence. J. Cell Sci. 111, 3129–3143 ( 1998).
Ruiz-Echevarria, M. J., Yasenchak, J. M., Han, X., Dinman, J. D. & Peltz, S. W. The upf3 protein is a component of the surveillance complex that monitors both translation and mRNA turnover and affects viral propagation. Proc. Natl Acad. Sci. USA 95, 8721–8726 (1998).
Henry, M., Borland, C. Z., Bossie, M. & Silver, P. A. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics 142, 103–115 ( 1996).
Xu, N., Chen, C. Y. & Shyu, A. B. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol. 17, 4611–4621 (1997).
Wagner, B. J., DeMaria, C. T., Sun, Y., Wilson, G. M. & Brewer, G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms . Genomics 48, 195–202 (1998).
Wilson, G. M., Sun, Y., Lu, H. & Brewer, G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J. Biol. Chem. 274, 33374–33381 (1999).
Ma, W. J., Cheng, S., Campbell, C., Wright, A. & Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271, 8144–8151 (1996).
Fan, X. C. & Steitz, J. A. Overexpression of HuR, a nuclear–cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17, 3448– 3460 (1998).
Fan, X. C. & Steitz, J. A. HNS, a nuclear–cytoplasmic shuttling sequence in HuR. Proc. Natl Acad. Sci. USA 95, 15293–15298 (1998).
Brennan, C. M., Gallouzi, I. E. & Steitz, J. A. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151, 1–14 (2000).
Gallouzi, I. E. et al. HuR binding to cytoplasmic mRNA is perturbed by heat shock . Proc. Natl Acad. Sci. USA 97, 3073– 3078 (2000).
Antic, D., Lu, N. & Keene, J. D. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 13, 449–461 ( 1999).
Gueydan, C. et al. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor-α mRNA. J. Biol. Chem. 274, 2322–2326 (1999).
Cunningham, K. S., Dodson, R. E., Nagel, M. A., Shapiro, D. J. & Schoenberg, D. R. Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′-untranslated region by the mRNA endonuclease polysomal ribonuclease 1. Proc. Natl Acad. Sci. USA 97, 12498–12502 (2000).
Author information
Authors and Affiliations
Supplementary information
Related links
Related links
DATABASE LINKS
FURTHER INFORMATION
ENCYCLOPEDIA OF LIFE SCIENCES
Glossary
- CAP ANALOGUES
-
Dinucleotides such as 7-meGpppG or GpppG that resemble the 5′ cap structure of messenger RNAs. They can be used to analyse the specificity of cap-binding proteins or to compete them away from the 5′ cap structure.
- 3′ UNTRANSLATED REGION
-
Non-coding region that lies 3′ to the protein-coding part of a messenger RNA. Often contains sequences involved in RNA regulation.
- 5′ UNTRANSLATED REGION
-
Non-coding region that lies in front of (5′ to) the protein-coding part of a messenger RNA.
- DOWNSTREAM SEQUENCE ELEMENT
-
A degenerate sequence element found in the coding region of most mRNAs, which can promote nonsense-mediated decay when located downstream of a premature stop codon.
- HETEROLOGOUS NUCLEAR RIBONUCLEOPROTEIN
-
RNA-binding protein with a nuclear function. Many hnRNPs can shuttle between the nucleus and the cytoplasm, indicating that they might function in nuclear export of RNA.
- MS2 COAT PROTEIN
-
A specific mRNA-binding protein from bacteriophage MS2 that recognizes a stem–loop structure. MS2 coat protein is often used to tether other proteins to RNAs.
- POLYSOME
-
Polyribosome; two or more ribosomes attached to different points on the same strand of mRNA.
Rights and permissions
About this article
Cite this article
Wilusz, C., Wormington, M. & Peltz, S. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2, 237–246 (2001). https://doi.org/10.1038/35067025
Issue Date:
DOI: https://doi.org/10.1038/35067025
This article is cited by
-
The multifaceted role of Fragile X-Related Protein 1 (FXR1) in cellular processes: an updated review on cancer and clinical applications
Cell Death & Disease (2024)
-
ReadZS detects cell type-specific and developmentally regulated RNA processing programs in single-cell RNA-seq
Genome Biology (2022)
-
Disease-associated human genetic variation through the lens of precursor and mature RNA structure
Human Genetics (2022)
-
Aging through an epitranscriptomic lens
Nature Aging (2021)
-
Igf2bp3 maintains maternal RNA stability and ensures early embryo development in zebrafish
Communications Biology (2020)