Abstract
Free genetic elements can be readily integrated into bacterial chromosomes, but so far, with the exception of one virus, there has been no evidence that this happens in Archaea — the other domain of microorganisms. Here we show that site-specific integration of different genetic elements into archaeal chromosomes is a general phenomenon, albeit rare, which requires an archaeal integrase and produces a partitioned integrase gene in the chromosome. The process is distinct from bacterial mechanisms and has implications for how horizontal gene transfer might occur across the boundaries of the domains of life.
Similar content being viewed by others
Main
Many bacterial integrases belong to a superfamily of tyrosine recombinase enzymes1 and mediate site-specific recombination between small extra-chromosomal DNA elements and chromosomes2, facilitating horizontal gene transfer between bacteria2. The only known archaeal integrase effects site-specific and reversible integration of the SSV1 virus into the chromosome of Sulfolobus shibatae3,4 (Fig. 1a, top).
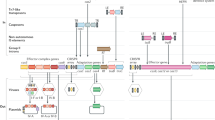
a, Organization of the SSV1 virus and the putative genetic elements pXQ1 and XQ2 in the chromosomes of S. shibatae and S. solfataricus. The relative positions of the partitioned integrase genes, the att sites (45 base pairs) and the overlapping tRNA genes are indicated. b, Alignment of the direct repeats attL and attR from virus SSV1 and from pXQ1 and XQ2. Nucleotides are shaded if two or more positions are identical. Arrows indicate a 5-base-pair inverted repeat.
We have previously identified a region of the Sulfolobus solfataricus genome that shows high sequence similarity to the Sulfolobus plasmid pHEN7 and to other members of the pRN plasmid family5,6,7. This chromosomal region is flanked by open-reading frames (ORFs) with extensive sequence similarity to the amino-terminal (66 amino acids) and carboxy-terminal (269 amino acids) regions of the SSV1 integrase (Fig. 1a). The former, Int(N), overlaps with the downstream half of a gene encoding tRNAVal and shows 71% sequence identity to the 45-base-pair target site (att) of the SSV1 integrase4 (within the downstream half of a tRNAArg gene), whereas the latter, Int(C), contains an identical att site (Fig. 1a, b).
Excision from the chromosome by recombination at the direct att repeat and circularization creates a plasmid, pXQ1, containing an intact integrase gene and one copy of the att site, similar to that of the SSV1 virus4. Thus, with the aid of the encoded integrase, pXQ1 could have integrated into the chromosome. As neither the free form of pXQ1 nor the empty chromosomal site of Sulfolobus were detectable by polymerase chain reaction (our unpublished results), we infer that the absence of an intact integrase gene precludes integrase-mediated excision from the chromosome.
A search for more homologues of the SSV1 integrase encoded in the chromosome of S. solfataricus5 revealed three Int(N)s and one Int(C). The sequences encoding Int(N) overlap with downstream halves of tRNA genes and all the sequences contain att sites. By analogy to the SSV1 virus, a genetic element XQ2 (67.7 kilobases) can be formed by recombination and excision at the attL and attR sites (Fig. 1a, b).
Four of the ORFs encoded in XQ2 were assigned to genes encoding the enzymes dTDP–glucose-4,6-dehydratase, glucose-1-phosphate–thymidylyl transferase, dTDP– 4-dehydrorhamnose reductase and dTDP– 4-dehydrorhamnose-3,5-epimerase, all of which are involved in central metabolism, and three of which are generally clustered in one operon. However, there are two additional copies of dTDP–glucose-4,6-dehydratase and another copy of each of the other enzymes encoded within the S. solfataricus chromosome5 that show 76–84% sequence identity and 86–96% similarity. It is therefore likely that XQ2 entered the chromosome, aided by its own integrase, to produce a gene-capture event.
We searched the complete genome sequences of seven other archaea for integrase homologues8. Those from the euryarchaeote Pyrococcus horikoshii9 and the crenarchaeote Aeropyrum pernix10 each had two Int(N), overlapping downstream halves of tRNA genes, and two and one copy of Int(C), respectively. The genome of Pyrococcus OT3 has two partitioned integrase genes11. Complementary fragments of the integrase genes contain att sites and border regions of 21.5 and 4 kilobases in P. horikoshii and 17.5 kilobases in A. pernix. Compatible with the idea that these are inserts from unknown organisms, none of their ORFs reveals any hits in the GenBank/EMBL databases8. We have detected a total of seven partitioned integrase genes, on average one per genome. Their sequences vary markedly in the amino-terminal region but are less variable in the carboxy-terminal region (22–33%/41–51% identity/ similarity), indicating that other integrase genes may still remain to be discovered.
We conclude that chromosomal integration in archaea differs from that in bacteria in producing partitioned integrase genes and that integration can be reversed only if the intact integrase is produced4. 'Curing' the cell of the free genetic element carrying the intact integrase gene can thus lead to gene capture by the archaeal chromosome. Although archaeal integrases differ from bacterial integrases in lacking a conserved motif1 and in having att sites in the coding region, they may still facilitate gene transfer between archaea, bacteria and eukaryotes12.
References
Nunes-Düby, S. E. et al. Nucleic Acids Res. 26, 391–406 (1998).
Rowe-Magnus, D. A. & Mazel, D. Curr. Opin. Microbiol. 2, 483–488 (1999).
Reiter, W.-D & Palm, P. Mol. Gen. Genet. 221, 65–71 (1990).
Muskhelishvili, G., Palm, P. & Zillig, W. Mol. Gen. Genet. 237, 334–342 (1993).
She, Q. et al. DNA Sequence 11, 183–192 (2000).
Kletzin, A., Lieke, A., Urich, T., Charlebois, R. L. & Sensen, C. W. Genetics 152, 1307–1314 (1999).
Peng, X., Holz, I., Zillig, W., Garrett, R. A. & She, Q. J. Mol. Biol. 303, 449–454 (2000).
Altschul, S. F. et al. Nucleic Acids Res. 25, 3389–3402 (1997).
Kawarabayasi, Y. et al. DNA Res. 5, 55–76 (1998).
Kawarabayasi, Y. et al. DNA Res. 6, 83–101 (1999).
Makino, S., Amano, N., Koike, H. & Suzuki, M. Proc. Jpn Acad. Ser. B 75, 166–171 (1999).
Groth, A. C., Olivares, E. C., Thyagarajan, B. & Calos, M. P. Proc. Natl Acad. Sci. USA 97, 5995–6000 (2000).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
She, Q., Peng, X., Zillig, W. et al. Gene capture in archaeal chromosomes. Nature 409, 478 (2001). https://doi.org/10.1038/35054138
Issue Date:
DOI: https://doi.org/10.1038/35054138
This article is cited by
-
Mechanisms of gene flow in archaea
Nature Reviews Microbiology (2017)
-
Molecular biology of fuselloviruses and their satellites
Extremophiles (2014)
-
Archaeal viruses—novel, diverse and enigmatic
Science China Life Sciences (2012)
-
Discovery of permuted and recently split transfer RNAs in Archaea
Genome Biology (2011)
-
Functional curation of the Sulfolobus solfataricus P2 and S. acidocaldarius 98-3 complete genome sequences
Extremophiles (2011)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.