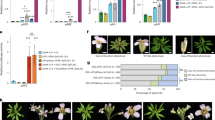
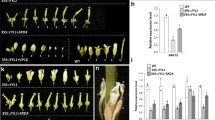
Abstract
Genetic studies, using floral homeotic mutants, have led to the ABC model of flower development. This model proposes that the combinatorial action of three sets of genes, the A, B and C function genes, specify the four floral organs (sepals, petals, stamens and carpels) in the concentric floral whorls1,2. However, attempts to convert vegetative organs into floral organs by altering the expression of ABC genes have been unsuccessful3,4,5. Here we show that the class B proteins of Arabidopsis, PISTILLATA (PI) and APETALA3 (AP3), interact with APETALA1 (AP1, a class A protein) and SEPALLATA3 (SEP3, previously AGL9), and with AGAMOUS (AG, a class C protein) through SEP3. We also show that vegetative leaves of triply transgenic plants, 35S::PI;35S::AP3;35S::AP1 or 35S::PI;35S::AP3;35S::SEP3, are transformed into petaloid organs and that those of 35S::PI; 35S::AP3;35S::SEP3;35S::AG are transformed into staminoid organs. Our findings indicate that the formation of ternary and quaternary complexes of ABC proteins may be the molecular basis of the ABC model, and that the flower-specific expression of SEP3 restricts the action of the ABC genes to the flower.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Coen, E. S. & Meyerowitz, E. M. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37 (1991).
Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20 (1991).
Mizukami, Y. & Ma, H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119-131 ( 1992).
Krizek, B. A. & Meyerowitz, E. M. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122, 11– 22 (1996).
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E. & Yanofsky, M. F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203 (2000).
Mandel, M. A., Gustafson-Brown, C., Savidge, B. & Yanofsky, M. F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273– 277 (1992).
Goto, K. & Meyerowitz, E. M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560 ( 1994).
Jack, T., Brockman, L. L. & Meyerowitz, E. M. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683-697 (1992).
Yanofsky, M. F. et al. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39 (1990).
Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H. & Sommer, H. Genetic control of flower development: homeotic genes in Antirrhinum majus. Science 250, 931–936 (1990).
Ma, H., Yanofsky, M. F. & Meyerowitz, E. M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5, 484–495 ( 1991).
Riechmann, J. L., Krizek, B. A. & Meyerowitz, E. M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl Acad. Sci. USA 93, 4793– 4798 (1996).
Herskowitz, I. A regulatory hierarchy for cell specialization in yeast. Nature 342, 749–757 ( 1989).
Tilly, J. J., Allen, D. W. & Jack, T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125, 1647– 1657 (1998).
Hill, T. A., Day, C. D., Zondlo, S. C., Thackeray, A. G. & Irish, V. F. Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125, 1711– 1721 (1998).
Honma, T. & Goto, K. The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 127, 2021–2030 (2000).
Sadowski, I., Ma, J., Triezenberg, S. & Ptashne, M. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335, 563–564 (1988).
Mandel, M. A. & Yanofsky, M. F. The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex Plant Reprod. 11, 22–28 (1998).
Rubinelli, P., Hu, Y. & Ma, H. Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 37, 607–619 (1998).
Fan, H. -Y., Hu, Y., Tudor, M. & Ma, H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 12, 999– 1010 (1997).
Cho, S. et al. Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol. Biol. 40, 419–429 ( 1999).
Riechmann, J. L. & Meyerowitz, E. M. MADS domain proteins in plant development. J. Biol. Chem. 378, 1079–1101 (1997).
Egea-Cortines, M., Saedler, H. & Sommer, H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379 (1999).
Davies, B., Egea-Cortines, M., de Andrade Silva, E., Saedler, H. & Sommer, H. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 15, 4330-4343 (1996).
Rounsley, S. D., Ditta, G. S. & Yanofsky, M. F. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7, 1259– 1269 (1995).
Smyth, D. A reverse trend—MADS functions revealed. Trends Plant Sci. 5, 315–317 ( 2000).
Parcy, F., Nilsson, O., Busch, M. A., Lee, I. & Weigel, D. A genetic framework for floral patterning. Nature 395, 561–566 (1998).
Bartel, P. L., Chien, C., Sternglanz, R. & Fields, S. in Cellular Interactions in Development: a Practical Approach. (ed. Hartley, D. A.) 153–179 (IRL Press, Oxford, 1993).
Shiraishi, H., Okada, K. & Shimura, Y. Nucleotide sequences recognized by the AGAMOUS MADS domain of Arabidopsis thaliana in vitro. Plant J. 4, 385–398 (1993).
Pan, S., Sehnke, P. C., Ferl, R. J. & Gurley, W. B. Specific interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex. Plant Cell 11, 1591– 1602 (1999).
Bechtold, N., Ellis, J. & Pelletier, G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis plants. C. R. Acad. Sci. Paris 316, 1194–1199 (1993).
Acknowledgements
We are grateful to M. Yanofsky for communicating data before publication, and to D. Weigel for providing the cDNA library. We also thank J. Bowman, T. Ito and H. Tsukaya for critical reading of the manuscript. This work was supported by grants from the Monbusho and JSPS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Institute for Chemical Research, Kyoto University, Uji, 611-0011, Japan
Rights and permissions
About this article
Cite this article
Honma, T., Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525–529 (2001). https://doi.org/10.1038/35054083
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35054083
This article is cited by
-
One pattern analysis (OPA) for the quantitative determination of protein interactions in plant cells
Plant Methods (2023)
-
Cloning and Expression Analysis of Onion (Allium cepa L.) MADS-Box Genes and Regulation Mechanism of Cytoplasmic Male Sterility
Biochemical Genetics (2023)
-
Dissecting SEPALLATA3 Splicing Variant Functions During Arabidopsis Vegetative Growth by amiRNA Technology
Journal of Plant Growth Regulation (2023)
-
Genome-wide characterization of Ficus carica MADS-box transcription factors with a focus on their roles during fruit development
Horticulture, Environment, and Biotechnology (2023)
-
Molecular mechanisms regulating ornamental traits and scent production in snapdragon (Antirrhinum majus L.)
Horticulture Advances (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.