Abstract
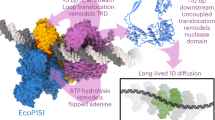
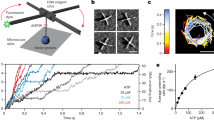
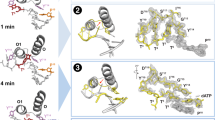
Helical filaments driven by linear molecular motors are anticipated to rotate around their axis, but rotation consistent with the helical pitch has not been observed. 14S dynein1 and non-claret disjunctional protein (ncd)2 rotated a microtubule more efficiently than expected for its helical pitch, and myosin rotated an actin filament only poorly3. For DNA-based motors such as RNA polymerase, transcription-induced supercoiling of DNA4 supports the general picture of tracking along the DNA helix5. Here we report direct and real-time optical microscopy measurements of rotation rate that are consistent with high-fidelity tracking. Single RNA polymerase molecules attached to a glass surface rotated DNA for >100 revolutions around the right-handed screw axis of the double helix with a rotary torque of >5 pN nm. This real-time observation of rotation opens the possibility of resolving individual transcription steps.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Vale, R. D. & Toyoshima, Y. Y. Rotation and translocation of microtubules in vitro induced by dyneins from Tetrahymena cilia. Cell 52, 459–469 (1988).
Walker, R. A., Salmon, E. D. & Endow, S. A. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature 347, 780–782 (1990).
Sase, I., Miyata, H., Ishiwata, S. & Kinosita, K. Jr Axial rotation of sliding actin filaments revealed by single-fluorophore imaging. Proc. Natl Acad. Sci. USA 94, 5646– 5650 (1997).
Wang, J. C. & Lynch, A. S. Transcription and DNA supercoiling. Curr. Opin. Genet. Dev. 3, 764– 768 (1993).
Cook, P. R. The organization of replication and transcription. Science 284, 1790–1795 (1999).
Schafer, D. A., Gelles, J., Sheetz, M. P. & Landick, R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature 352, 444–448 (1991).
Wang, M. D. et al. Force and velocity measured for single molecules of RNA polymerase. Science 282, 902–907 (1999).
Guthold, M. et al. Direct observation of one-dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys. J. 77, 2284–2294 (1999).
Davenport, R. J., Wuite, G. J. L., Landick, R. & Bustamante, C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science 287, 2497– 2500 (2000).
Kabata, H. et al. Visualization of single molecules of RNA polymerase sliding along DNA. Science 262, 1561– 1563 (1993).
Harada, Y. et al. Single molecule imaging of RNA polymerase-DNA interactions in real time. Biophys. J. 76, 709– 715 (1999).
Levin, J. R., Krummel, B. & Chamberlin, M. J. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at single template base. J. Mol. Biol. 196, 85– 100 (1987).
Stryer, L. Biochemistry 4th edn (Freeman, New York, 1995).
Svoboda, K. & Block, S. M. Biological applications of optical tweezers. Annu. Rev. Biophys. Biomol. Struct. 23, 247–285 (1994).
Strick, T., Allemand, J.-F., Bensimon, D., Lavery, R. & Croquette, V. Phase coexistence in a single DNA molecule. Physica A 263, 392– 405 (1999).
Noji, H., Yasuda, R., Yoshida, M. & Kinosita, K. Jr Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 ( 1997).
Yasuda, R., Noji, H., Kinosita, K. Jr & Yoshida, M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell 93, 1117– 1124 (1998).
DeRosier, D. J. The turn of the screw: the bacterial flagellar motor. Cell 93, 17–20 (1998).
Ryu, W. S., Berry, R. M. & Berg, H. C. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403, 444–447 (2000).
Adachi, K. et al. Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl Acad. Sci. USA 97, 7243–7247 ( 2000).
Kubori, T. & Shimamoto, N. A branched pathway in the early stage of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 256, 449–457 (1996).
Studier, F. W. Gene 0·3 of bacteriophage T7 acts to overcome the DNA restriction system of the host. J. Mol. Biol. 94, 283–295 (1975).
Acknowledgements
We thank M. Susa for help in transcription analysis; A. Ishihama, S. Ishiwata, G. W. Feigenson and members of Team 13 for comments; and H. Umezawa for laboratory management. This work was supported in part by Grants-in-Aid from Ministry of Education, Science, Sports and Culture of Japan, Hayashi Memorial Foundation for Female Natural Scientists, and an Academic Frontier Promotional Project.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
41586_2001_BF35051126_MOESM1_ESM.doc
Analyses of transcription rates in solution (estimation by gel electrophoresis) and on individual RNAP molecules attached to a glass surface (estimation from the reduction in the range of brownian motion of the end bead).
Rights and permissions
About this article
Cite this article
Harada, Y., Ohara, O., Takatsuki, A. et al. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature 409, 113–115 (2001). https://doi.org/10.1038/35051126
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35051126
This article is cited by
-
A genetic circuit on a single DNA molecule as an autonomous dissipative nanodevice
Nature Communications (2024)
-
Ideas and methods of nonlinear mathematics and theoretical physics in DNA science: the McLaughlin-Scott equation and its application to study the DNA open state dynamics
Biophysical Reviews (2021)
-
DNA:RNA hybrid G-quadruplex formation upstream of transcription start site
Scientific Reports (2020)
-
Rotation tracking of genome-processing enzymes using DNA origami rotors
Nature (2019)
-
Theoretical Model of Transcription Based on Torsional Mechanics of DNA Template
Journal of Statistical Physics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.