Abstract
Splicing of pre-messenger RNA is regulated differently in the brain compared with other tissues. Recognition of aberrations in splicing events that are associated with neurological disease has contributed to our understanding of disease pathogenesis in some cases. Neuron-specific proteins involved in RNA splicing and metabolism are also affected in several neurological disorders. These findings have begun to bridge what we know about the mechanisms regulating neuron-specific splicing and our understanding of neural function and disease.
Key Points
-
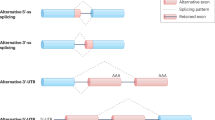
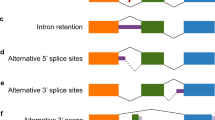
Alternative splicing is the process by which different proteins are produced from a single pre-messenger RNA by regulating the choice of exons to be included in the mature transcript. Alternative splicing involves a group of proteins responsible for the splicing event itself, as well as other molecules important for its regulation.
-
In the nervous system, different disorders are related to abnormal alternative splicing. The two best characterized are inherited frontotemporal dementia and parkinsonism linked to chromosome 17, and spinal muscular atrophy. In both cases, splicing defects of identified exons have been associated to the disease phenotype. Other neurological conditions, such as spinocerebellar ataxia 8 and amyotrophic lateral sclerosis, are also thought to relate to abnormal splicing, but the evidence is still incomplete.
-
Although splicing events in the nervous system involve similar factors and regulatory elements as splicing in other cell types, the study of the paraneoplastic neurological disorders has led to the identification of brain-specific factors that regulate alternative splicing. For instance, NOVA-1, a molecule linked to paraneoplastic opsoclonus myoclonus ataxia, is involved in splicing of glycine and GABA (γ-aminobutyric acid) receptor subunits.
-
How could neuron-specific splicing be regulated? It has been proposed that neuron-specific splicing is selectively repressed in other tissues by trans-acting RNA-binding proteins. This repression would be relieved in neurons by the presence of positive regulators that compete for the same binding sites on the pre-mRNA. Alternatively, the presence or absence of regulatory factors, or of specific binding sites within the mRNA, could permit splicing events to be regulated independently of each other within different cells at different times.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Lin, Z., Haus, S., Edgerton, J. & Lipscombe, D. Identification of functionally distinct isoforms of the N-type Ca2+ channel in rat sympathetic ganglia and brain. Neuron 18, 153–166 (1997).
Ehlers, M. D., Tingley, W. G. & Huganir, R. L. Regulated subcellular distribution of the NR1 subunit of the NMDA receptor. Science 269, 1734– 1737 (1995).
Macdonald, R. L. Ethanol, γ-aminobutyrate type A receptors, and protein kinase C phosphorylation . Proc. Natl Acad. Sci. USA 92, 3633– 3635 (1995).
Picetti, R. et al. Dopamine D2 receptors in signal transduction and behavior . Crit. Rev. Neurobiol. 11, 121– 142 (1997).
Conn, P. J. & Pin, J. P. Pharmacology and functions of metabotropic glutamate receptors. Annu. Rev. Pharmacol. Toxicol. 37, 205–237 (1997).
Stamm, S., Zhang, M. Q., Marr, T. G. & Helfman, D. M. A sequence compilation and comparison of exons that are alternatively spliced in neurons. Nucleic Acids Res. 22, 1515– 1526 (1994).
Amara, S. G., Jonas, V., Rosenfeld, M. G., Ong, E. S. & Evans, R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 298, 240–244 ( 1982).
Buckanovich, R. J., Posner, J. B. & Darnell, R. B. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron 11, 657– 672 (1993).Cloning and identification of NOVA-1 as the target antigen in paraneoplastic opsoclonus myoclonus ataxia. NOVA-1 is a novel brain-specific protein with homology to RNA-binding proteins, indicating a possible role in the regulation of RNA splicing or metabolism specific to a subset of neurons.
Jensen, K. B. et al. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25, 359–371 (2000).Biochemical studies show that NOVA-1 regulates alternative splicing of two inhibitory transmitter receptor subunit pre-mRNAs, a finding confirmed in Nova-1 -null mice.
Hutton, M. et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705 (1998). Six independent mutations within the tau gene are associated with FTDP-17. Three are intronic mutations at the 5′ splice site, which result in an increase in the incorporation of exon 10.
Spillantini, M. G. et al. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl Acad. Sci. USA 95, 7737–7741 (1998).
Hong, M. et al. Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science 282, 1914 –1917 (1998).
Poorkaj, P. et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia . Ann. Neurol. 43, 815– 825 (1998); erratum 44, 428 (1998).
Hasegawa, M., Smith, M. J. & Goedert, M. Tau proteins with FTDP-17 mutations have a reduced ability to promote microtubule assembly. FEBS Lett. 437, 207–210 (1998).
D'Souza, I. et al. Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc. Natl Acad. Sci. USA 96, 5598–5603 (1999).
Goedert, M. & Spillantini, M. G. Tau mutations in frontotemporal dementia FTDP-17 and their relevance for Alzheimer's disease. Biochim. Biophys. Acta 1502, 110–121 (2000).
Goedert, M., Spillantini, M. G., Potier, M. C., Ulrich, J. & Crowther, R. A. Cloning and sequencing of the cDNA encoding an isoform of microtubule- associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J. 8, 393–399 (1989).
Kosik, K. S., Orecchio, L. D., Bakalis, S. & Neve, R. L. Developmentally regulated expression of specific tau sequences. Neuron 2, 1389–1397 ( 1989).
Grover, A. et al. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 274, 15134–15143 (1999).
Jiang, Z., Cote, J., Kwon, J. M., Goate, A. M. & Wu, J. Y. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell. Biol. 20, 4036–4048 (2000).
Ishihara, T. et al. Age-dependent emergence and progression of a tauopathy in transgenic mice overexpressing the shortest human tau isoform. Neuron 24, 751–762 ( 1999).
Brzustowicz, L. M. et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2–13.3. Nature 344 , 540–541 (1990).
Melki, J. et al. Gene for chronic proximal spinal muscular atrophies maps to chromosome 5q. Nature 344, 767–768 (1990).
Lefebvre, S. et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165 (1995).
Liu, Q. & Dreyfuss, G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 15, 3555–3565 (1996).
Pellizzoni, L., Kataoka, N., Charroux, B. & Dreyfuss, G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95, 615– 624 (1998).
Pellizzoni, L., Charroux, B. & Dreyfuss, G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl Acad. Sci. USA 96, 11167–11172 ( 1999).
Lorson, C. L. et al. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nature Genet. 19, 63– 66 (1998).
Lorson, C. L., Hahnen, E., Androphy, E. J. & Wirth, B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA 96, 6307–6311 (1999). Systematic study of the nucleotides that differ between SMN1 and SMN2 , which shows that a single nucleotide exchange attenuates the activity of an exonic splicing enhancer.
Lorson, C. L. & Androphy, E. J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 9, 259–265 (2000).
Brown, R. H. Jr Superoxide dismutase and familial amyotrophic lateral sclerosis: new insights into mechanisms and treatments. Ann. Neurol. 39, 145–146 (1996).
Siddique, T. & Deng, H. X. Genetics of amyotrophic lateral sclerosis. Hum. Mol. Genet. 5, 1465– 1470 (1996).
Rothstein, J. D., Martin, L. J. & Kuncl, R. W. Decreased glutamate transport by the brain and spinal cord in amyotrophic lateral sclerosis. N. Engl. J. Med. 326, 1464–1468 (1992).
Rothstein, J. D. et al. Localization of neuronal and glial glutamate transporters . Neuron 13, 713–725 (1994).
Shaw, P. J., Forrest, V., Ince, P. G., Richardson, J. P. & Wastell, H. J. CSF and plasma amino acid levels in motor neuron disease: elevation of CSF glutamate in a subset of patients. Neurodegeneration 4, 209–216 ( 1995).
Lin, C. L. et al. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis . Neuron 20, 589–602 (1998).
Meyer, T. et al. The RNA of the glutamate transporter EAAT2 is variably spliced in amyotrophic lateral sclerosis and normal individuals. J. Neurol. Sci. 170, 45–50 ( 1999).
Honig, L. S., Chambliss, D. D., Bigio, E. H., Carroll, S. L. & Elliott, J. L. Glutamate transporter EAAT2 splice variants occur not only in ALS, but also in AD and controls. Neurology 55, 1082–1088 ( 2000).
Koob, M. D. et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nature Genet. 21, 379– 384 (1999).A non-coding CTG expansion causes a novel form of spinocerebellar ataxia (SCA), the first example of a dominant SCA not caused by a polyglutamine expansion.
Taneja, K. L., McCurrach, M., Schalling, M., Housman, D. & Singer, R. H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 128, 995–1002 ( 1995).
Davis, B. M., McCurrach, M. E., Taneja, K. L., Singer, R. H. & Housman, D. E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl Acad. Sci. USA 94, 7388–7393 (1997).
Mankodi, A. et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 289, 1769– 1773 (2000).
Timchenko, L. T. et al. Identification of a (CUG)– triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 24, 4407–4414 ( 1996).
Philips, A. V., Timchenko, L. T. & Cooper, T. A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280, 737–741 (1998).Evidence that CUG repeats associated with myotonic dystrophy might disrupt regulation of alternative splicing through interactions with CUG-binding protein.
Darnell, R. B. Onconeural antigens and the paraneoplastic neurologic disorders: at the intersection of cancer, immunity, and the brain. Proc. Natl Acad. Sci. USA 93, 4529–4536 (1996).
Musunuru, K. & Darnell, R. B. Paraneoplastic neurologic disease antigens: RNA-binding proteins and signaling proteins in neuronal degeneration . Annu. Rev. Neurosci. (in the press).
Yang, Y. Y., Yin, G. L. & Darnell, R. B. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc. Natl Acad. Sci. USA 95, 13254–13259 (1998).
Ostareck, D. H. et al. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89, 597–606 ( 1997).
Kiledjian, M., Wang, X. & Liebhaber, S. A. Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J. 14, 4357 –4364 (1995).
Deshler, J. O., Highett, M. I., Abramson, T. & Schnapp, B. J. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 8, 489– 496 (1998).
Ross, A. F., Oleynikov, Y., Kislauskis, E. H., Taneja, K. L. & Singer, R. H. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17, 2158–2165 (1997).
Pieretti, M. et al. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66, 817– 822 (1991).
De Boulle, K. et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nature Genet. 3, 31–35 (1993).
Min, H., Turck, C. W., Nikolic, J. M. & Black, D. L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11, 1023– 1036 (1997).
Arning, S., Gruter, P., Bilbe, G. & Kramer, A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA 2, 794–810 (1996).
Siebel, C. W., Admon, A. & Rio, D. C. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev. 9, 269–283 (1995).
Engebrecht, J. A., Voelkel-Meiman, K. & Roeder, G. S. Meiosis-specific RNA splicing in yeast. Cell 66, 1257–1268 ( 1991).
Buckanovich, R. J. & Darnell, R. B. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 17, 3194–3201 (1997).
Jensen, K. B., Musunuru, K., Lewis, H. A., Burley, S. K. & Darnell, R. B. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl Acad. Sci. USA 97, 5740– 5755 (2000).
Lewis, H. A. et al. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100, 323–332 (2000).
Grabowski, P. J. Genetic evidence for a Nova regulator of alternative splicing in the brain . Neuron 25, 254–256 (2000).
Dalmau, J., Graus, F., Rosenblum, M. K. & Posner, J. B. Anti-Hu-associated paraneoplastic encephalomyelitis/sensory neuronopathy. A clinical study of 71 patients. Medicine (Baltimore) 71, 59–72 (1992).
Graus, F., Cordon-Cardo, C. & Posner, J. B. Neuronal antinuclear antibody in sensory neuronopathy from lung cancer. Neurology 35, 538– 543 (1985).
Dalmau, J., Furneaux, H. M., Cordon-Cardo, C. & Posner, J. B. The expression of the Hu (paraneoplastic encephalomyelitis/sensory neuronopathy) antigen in human normal and tumor tissues. Am. J. Pathol. 141, 881–886 (1992).
Okano, H. J. & Darnell, R. B. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 17, 3024–3037 (1997).
Szabo, A. et al. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell 67, 325–333 (1991).
Keene, J. D. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl Acad. Sci. USA 96, 5–7 (1999).
Brennan, C. M. & Steitz, J. A. HuR and mRNA stability. Cell. Mol. Life Sci. (in the press).
Robinow, S., Campos, A. R., Yao, K. M. & White, K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science 242, 1570– 1572 (1988).
Koushika, S. P., Lisbin, M. J. & White, K. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr. Biol. 6, 1634–1641 (1996).
Koushika, S. P., Soller, M. & White, K. The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol. Cell. Biol. 20, 1836–1845 (2000).
Manley, G. T., Smitt, P. S., Dalmau, J. & Posner, J. B. Hu antigens: reactivity with Hu antibodies, tumor expression, and major immunogenic sites . Ann. Neurol. 38, 102– 110 (1995).
Zhang, L., Ashiya, M., Sherman, T. G. & Grabowski, P. J. Essential nucleotides direct neuron-specific splicing of γ-2 pre-mRNA . RNA 2, 682–698 (1996).
Wang, Z. & Grabowski, P. J. Cell- and stage-specific splicing events resolved in specialized neurons of the rat cerebellum. RNA 2, 1241–1253 ( 1996).
Ashiya, M. & Grabowski, P. J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA 3, 996– 1015 (1997).Evidence that polypyrimidine-tract-binding protein represses neuron-specific splicing of GABA A γ2 pre-mRNA in non-neuronal cells.
Zhang, L., Liu, W. & Grabowski, P. J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA 5, 117–130 (1999).
Chan, R. C. & Black, D. L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 17, 4667–4676 (1997). The polypyrimidine-tract-binding protein represses neuron-specific splicing of c-src through interaction with an upstream sequence element in non-neuronal cells.
Chan, R. C. & Black, D. L. Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro. Mol. Cell. Biol. 15, 6377–6385 (1995).
Grabowski, P. J. Splicing regulation in neurons: tinkering with cell-specific control. Cell 92, 709–712 ( 1998).
Chou, M. Y., Rooke, N., Turck, C. W. & Black, D. L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 19, 69–77 (1999).
Polydorides, A. D., Okano, H. J., Yang, Y. Y., Stefani, G. & Darnell, R. B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of nova to regulate neuron-specific alternative splicing. Proc. Natl Acad. Sci. USA 97, 6350–6355 (2000). Cloning and identification of brain-enriched polypyrimidine-tract-binding protein and evidence for a role in the regulation of alternative splicing in neurons through interactions with NOVA.
Vezzani, A., Speciale, C., Della Vedova, F., Tamburin, M. & Benatti, L. Alternative splicing at the C-terminal but not at the N-terminal domain of the NMDA receptor NR1 is altered in the kindled hippocampus. Eur. J. Neurosci. 7, 2513–2517 (1995).
Daoud, R., Da Penha Berzaghi, M., Siedler, F., Hubener, M. & Stamm, S. Activity-dependent regulation of alternative splicing patterns in the rat brain. Eur. J. Neurosci. 11, 788–802 (1999).
Okabe, S., Miwa, A. & Okado, H. Alternative splicing of the C-terminal domain regulates cell surface expression of the NMDA receptor NR1 subunit. J. Neurosci. 19, 7781–7792 (1999).
Tingley, W. G., Roche, K. W., Thompson, A. K. & Huganir, R. L. Regulation of NMDA receptor phosphorylation by alternative splicing of the C-terminal domain. Nature 364, 70– 73 (1993).
Kanopka, A. et al. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393, 185– 187 (1998).
Cao, W., Jamison, S. F. & Garcia-Blanco, M. A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 3, 1456–1467 (1997).
Xiao, S. H. & Manley, J. L. Phosphorylation of the ASF/SF2 RS domain affects both protein–protein and protein–RNA interactions and is necessary for splicing. Genes Dev. 11, 334–344 (1997).
Petersen-Mahrt, S. K. et al. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 18, 1014–1024 ( 1999).
Missler, M. & Südhof, T. C. Neurexins: three genes and 1001 products. Trends Genet. 14, 20– 26 (1998).
Schmucker, D. et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671–684 (2000).
Kohmura, N. et al. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 20, 1137–1151 (1998).
Wu, Q. & Maniatis, T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97, 779–790 ( 1999).
Sugino, H. et al. Genomic organization of the family of CNR cadherin genes in mice and humans. Genomics 63, 75– 87 (2000).
Smith, C. W. & Valcarcel, J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25, 381–388 (2000).
Hertel, K. J., Lynch, K. W. & Maniatis, T. Common themes in the function of transcription and splicing enhancers. Curr. Opin. Cell Biol. 9, 350 –357 (1997).
Staley, J. P. & Guthrie, C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92, 315–326 (1998).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Glossary
- TAU
-
A neuronal protein that binds to microtubules, promoting their assembly and stability.
- MISSENSE MUTATION
-
A mutation in which an incorrect amino acid is incorporated into the protein.
- SILENT MUTATION
-
A mutation that is not accompanied by an amino-acid change in the translation product.
- EXON TRAPPING
-
A method for finding expressed DNA sequences that is based on the selection of functional splice sites in genomic DNA.
- MINIGENE
-
Sequence that contains all of the elements — such as the alternative exons and the surrounding introns — necessary to show the same splicing pattern as the endogenous gene.
- STEM-LOOP
-
A structure formed when a single-stranded DNA or RNA molecule loops back on itself to form a complementary double strand crowned by a loop.
- CIS-ACTING ELEMENT
-
A regulatory genetic element located in the same DNA molecule as the gene being regulated.
- LINKAGE MAPPING
-
The establishment of the position of a chromosomal locus based on the frequency of recombination.
- DOMINANT-NEGATIVE PROTEIN
-
A mutant molecule that forms heteromeric complexes with the wild type to yield a non-functional product.
- AUTOSOMAL
-
Related to any chromosome except the sex chromosomes.
- MYOTONIC DYSTROPHY
-
Inherited disorder characterized by delayed muscle relaxation after contraction. It is also accompanied by muscle weakness and wasting, and it can be associated with mild mental retardation, hair loss and cataracts.
- TROPONIN
-
A regulatory protein of striated muscle contraction. The T subunit binds to tropomyosin.
- EXPRESSION CLONING
-
Cloning strategy that is based on the transfection of complementary DNAs such that functional proteins are expressed, followed by a screening of the functional activity of the gene of interest.
- FRAGILE-X SYNDROME
-
A genetic condition commonly transmitted from mother to son, associated with mental retardation, abnormal facial features and enlarged testicles.
- LYMPHOKINES
-
Non-immunoglobulin molecules released by lymphocytes after a second interaction with a given antigen. Lymphokines are particularly important during cell-mediated immune reactions, as they affect the behaviour of other immune cell types.
- TRANS-ACTING ELEMENT
-
A regulatory genetic element whose effects are independent of its position and, therefore, can be located in a different DNA molecule to the gene being regulated.
- C-SRC
-
The first proto-oncogene to be identified. It codes for a non-receptor protein tyrosine kinase.
- CLATHRIN
-
One of the main protein components of the coats formed during membrane endocytosis.
- KINDLING
-
An experimental model of epilepsy in which an increased susceptibility to seizures arises after daily focal stimulation of specific brain areas (for example, the amygdala), stimulation that does not reach the threshold to elicit a seizure by itself.
- NEUREXINS
-
Family of membrane proteins implicated in axon guidance and synaptogenesis.
- CADHERINS
-
Calcium-dependent cell adhesion molecules that tend to engage in homophilic interactions.
Rights and permissions
About this article
Cite this article
Dredge, B., Polydorides, A. & Darnell, R. The splice of life: Alternative splicing and neurological disease . Nat Rev Neurosci 2, 43–50 (2001). https://doi.org/10.1038/35049061
Issue Date:
DOI: https://doi.org/10.1038/35049061
This article is cited by
-
Expression of a human cDNA in moss results in spliced mRNAs and fragmentary protein isoforms
Communications Biology (2021)
-
Alternative splicing and cancer: a systematic review
Signal Transduction and Targeted Therapy (2021)
-
Accumulation of poly(A) RNA in nuclear granules enriched in Sam68 in motor neurons from the SMNΔ7 mouse model of SMA
Scientific Reports (2018)
-
Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility
Nature Genetics (2018)
-
Alternative splicing isoforms in health and disease
Pflügers Archiv - European Journal of Physiology (2018)