Abstract
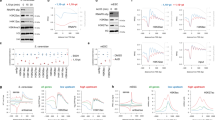
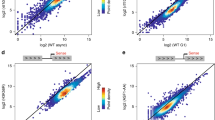
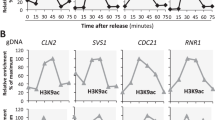
Histone acetyltransferases and deacetylases can be targeted to promoters to activate or repress genes. For example, the histone acetyltransferase GCN5 is part of a yeast multiprotein complex that is recruited by the DNA-binding activator protein GCN4 (refs 1, 2,3). The histone deacetylase RPD3 complex is recruited to DNA by the repressor UME6 (refs 4, 5); similar mechanisms exist in other eukaryotes6. However, deletion of RPD3 also increases expression of the PHO5 gene7 that is repressed by nucleosomes8,9, and regulated by GCN5 (ref. 10) but not by UME6. We have determined whether acetylation and deacetylation are promoter specific at PHO5, by using antibodies against acetylated lysine residues and chromatin immunoprecipitation to examine the acetylation state of a 4.25-kilobase region surrounding the PHO5 gene. Here we show that this region is acetylated extensively by ESA1 and GCN5 and deacetylated by HDA1 and RPD3, and that widespread histone modification affects three separate chromosomal regions examined, which total 22 kb. Our data indicate that targeted modification occurs in a background of global acetylation and deacetylation that not only reduces basal transcription, but also allows a rapid return to the initial state of acetylation when targeting is removed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Georgakopoulos, T. & Thireos, G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 11, 4145–4152 (1992).
Brownell, J. E. et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851 ( 1996).
Drysdale, C. M. et al. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol. Cell. Biol. 18, 1711– 1724 (1998).
Kadosh, D. & Struhl, K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18, 5121–5127 (1998).
Rundlett, S. E., Carmen, A. A., Suka, N., Turner, B. M. & Grunstein, M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392, 831–835 (1998).
Knoepfler, P. S. & Eisenman, R. N. Sin meets NuRD and other tails of repression. Cell 99, 447–450 (1999).
Vidal, M. & Gaber, R. F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 6317 –6327 (1991).
Svaren, J. & Hörz, W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22, 93–97 ( 1997).
Han, M. & Grunstein, M. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55, 1137–1145 (1988).
Gregory, P. D. et al. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell 1, 495–505 (1998).
Solomon, M. J. & Varshavsky, A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl Acad. Sci. USA 82, 6470– 6474 (1985).
Orlando, V., Strutt, H. & Paro, R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11, 205– 214 (1997).
Braunstein, M., Rose, A. B., Holmes, S. G., Allis, C. D. & Broach, J. R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7, 592–604 (1993).
Dedon, P. C., Soults, J. A., Allis, C. D. & Gorovsky, M. A. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal. Biochem. 197, 83–90 (1991).
Hecht, A., Strahl-Bolsinger, S. & Grunstein, M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383, 92–96 (1996).
Kuo, M. H., Zhou, J., Jambeck, P., Churchill, M. E. & Allis, C. D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12, 627–639 ( 1998).
Rundlett, S. E. et al. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA 93, 14503–14508 (1996).
Grant, P. A. et al. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895– 5900 (1999).
Clarke, A. S., Lowell, J. E., Jacobson, S. J. & Pillus, L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19, 2515– 2526 (1999).
Wittschieben, B. O. et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4, 123–128 (1999).
Reifnyder, C., Lowell, J., Clarke, A. & Pillus, L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nature Genet. 16, 109 (1997).
Ehrenhofer-Murray, A. E., Rivier, D. H. & Rine, J. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics 145, 923–934 ( 1997).
Parthun, M. R., Widom, J. & Gottschling, D. E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87, 85–94 ( 1996).
Hughes, T. R. et al. Functional discovery via a compendium of expression profiles. Cell 102, 109–126 (2000).
Han, M., Kim, U. J., Kayne, P. & Grunstein, M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae . EMBO J. 7, 2221–2228 (1988).
Schmitt, M. E., Brown, T. A. & Trumpower, B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18, 3091–3092 (1990).
Hecht, A. & Grunstein, M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 304, 399– 414 (1999).
Almer, A., Rudolph, H., Hinnen, A. & Hörz, W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5, 2689–2696 (1986).
Laman, H., Balderes, D. & Shore, D. Disturbance of normal cell cycle progression enhances the establishment of transcriptional silencing in Saccharomyces cerevisiae . Mol. Cell. Biol. 15, 3608– 3617 (1995).
Acknowledgements
We thank L. Pillus for the esa1ts strain. We are grateful to the members of the Grunstein laboratory for critical comments and discussions throughout this work and are thankful to Y. Suka and A. Carmen for antibodies against histone sites of acetylation. M.V. acknowledges pre-doctoral support from the Dottorato di Ricerca in Genetica e Biologia Molecolare—Università degli Studi di Roma “la Sapienza”. This work was supported by a Public Health Service grant from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vogelauer, M., Wu, J., Suka, N. et al. Global histone acetylation and deacetylation in yeast. Nature 408, 495–498 (2000). https://doi.org/10.1038/35044127
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35044127
This article is cited by
-
The histone deacetylase HOS2 controls pathogenicity through regulation of melanin biosynthesis and appressorium formation in Colletotrichum gloeosporioides
Phytopathology Research (2022)
-
Maternal Separation-Induced Histone Acetylation Correlates with BDNF-Programmed Synaptic Changes in an Animal Model of PTSD with Sex Differences
Molecular Neurobiology (2021)
-
Epigenetic regulation of matrix metalloproteinases in inflammatory diseases: a narrative review
Cell & Bioscience (2020)
-
A short guide to histone deacetylases including recent progress on class II enzymes
Experimental & Molecular Medicine (2020)
-
Disruption in phosphate transport affects membrane lipid and lipid droplet homeostasis in Saccharomyces cerevisiae
Journal of Bioenergetics and Biomembranes (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.