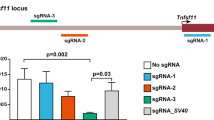
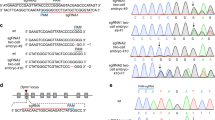
Abstract
GENE targeting in embryonic stem (ES) cells is a powerful tool for generating mice with null alleles1. Current methods of gene inactivation in ES cells introduce a neomycin gene (neo) cassette both as a mutagen and a selection marker for transfected cells2–11. Although null alleles are valuable, changes at the nucleotide level of a gene are very important for functional analysis. One gene family in which subtle mutations would be particularly valuable are the clusters of Hox homeobox genes12–16. Inactivation of genes in a cluster with a neo cassette that includes promoter/enhancer elements may deregulate transcription of neighbouring genes and generate a phenotype which is difficult to interpret. We describe here a highly efficient gene targeting method, termed the 'hit and run' procedure. This generates ES cells with subtle site-specific mutations with no selectable marker and may be useful for most genes. We have developed this procedure at the hypoxanthine phosphoribosyltransferase (hprt) locus and subsequently isolated ES cells with a premature stop codon in the homeobox of Hox-2.6 (ref. 14).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Capecchi, M. R. Science 244, 1288–1292 (1989).
Schwartzberg, P. L., Goff, S. P. & Robertson, E. J. Science 246, 799–803 (1989).
Zijlstra, M., Li, E., Sajjadi, F., Subramani, S. & Jaeniscu, R. Nature 342, 435–438 (1989).
McMahon, A. P. & Bradley, A. Cell 62, 1073–1085 (1990).
DeChiara, T. M., Efstratiadis, A. & Robertson, E. J. Nature 345, 78–80 (1990).
Koller, B. H. & Smithies, O. Science 248, 1227–1230 (1990).
Zijlstra, M. et al. Nature 344, 742–746 (1990).
Thomas, K. R. & Capecchi, M. R. Cell 51, 503–512 (1987).
Mansour, S. L., Thomas, K. R. & Capecchi, M. R. Nature 336, 348–352 (1988).
Johnson, R. S. et al. Science 245, 1234–1236 (1989).
Joyner, A. L., Skarnes, W. C. & Rossant, J. Nature 338, 153–156 (1989).
Graham, A., Papalopulu, N. & Krumlauf, R. Cell 57, 367–378 (1989).
Duboule, D. & Dolle, P. EMBO 8, 1497–1505 (1989).
Graham, A. et al. Genes Dev. 2, 1424–1438 (1988).
Gaunt, S. J., Krumlauf, R. & Doboule, D. Development 107, 131–142 (1989).
Wright, C. V. E. et al. Trends biochem. Sci. 14, 52–56 (1989).
Smithies, O., Gregg, R. G., Boggs, S. S., Koralewski, M. A. & Kucherlapati, R. S. Nature 317, 230–234 (1985).
Doetschman, T. et al. Nature 330, 576–578 (1987).
Baker, M. D. et al. Proc. natn. Acad. Sci. U.S.A. 85, 6432–6436 (1988).
Baker, M. D., Pennell, N., Bosnoyan, L. & Schulman, M. J. Molec. cell. Biol. 9, 5500–5507 (1989).
Borrelli, E., Heyman, R., Hsi, M. A. & Evans, R. M. Proc. natn. Acad. Sci. U.S.A. 85, 7572–7576 (1988).
Melton, D. W., Konecki, D. S., Brennand, J. & Caskey, C. T. Proc. natn. Acad. Sci. U.S.A. 81, 2147–2151 (1984).
Qian, Y. Q. et al. Cell 59, 573–580 (1989).
Zimmer, A. & Gruss, P. Nature 338, 150–153 (1989).
Holliday, R. Genet Res. 5, 282–304 (1964).
Adra, C. N., Boer, P. H. & McBurney, M. W. Gene 60, 65–74 (1987).
Pfarr, D. S. et al. DNA 5, 115–122 (1986).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hasty, P., Ramírez-Solis, R., Krumlauf, R. et al. Introduction of a subtle mutation into the Hox-2.6 locus in embryonic stem cells. Nature 350, 243–246 (1991). https://doi.org/10.1038/350243a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/350243a0
This article is cited by
-
Efficient simultaneous double DNA knock-in in murine embryonic stem cells by CRISPR/Cas9 ribonucleoprotein-mediated circular plasmid targeting for generating gene-manipulated mice
Scientific Reports (2022)
-
Progress and prospects: oligonucleotide-directed gene modification in mouse embryonic stem cells: a route to therapeutic application
Gene Therapy (2011)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.