Abstract
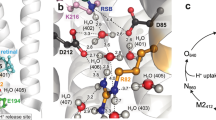
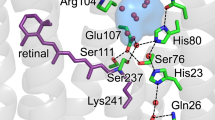
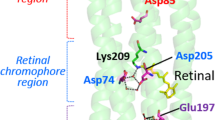
The transport of protons across membranes is an important process in cellular bioenergetics. The light-driven proton pump bacteriorhodopsin is the best-characterized protein providing this function. Photon energy is absorbed by the chromophore retinal, covalently bound to Lys 216 via a protonated Schiff base. The light-induced all-trans to 13-cis isomerization of the retinal results in deprotonation of the Schiff base followed by alterations in protonatable groups within bacteriorhodopsin. The changed force field induces changes, even in the tertiary structure1,2,3, which are necessary for proton pumping. The recent report4 of a high-resolution X-ray crystal structure for the late M intermediate of a mutant bacteriorhopsin (with Asp 96→Asn) displays the structure of a proton pathway highly disturbed by the mutation. To observe an unperturbed proton pathway, we determined the structure of the late M intermediate of wild-type bacteriorhodopsin (2.25 Å resolution). The cytoplasmic side of our M2 structure shows a water net that allows proton transfer from the proton donor group Asp 96 towards the Schiff base. An enlarged cavity system above Asp 96 is observed, which facilitates the de- and reprotonation of this group by fluctuating water molecules in the last part of the cycle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dencher, N. A., Dresselhaus, D., Zaccai, G. & Büldt, G. Structural changes in bacteriorhodopsin during proton translocation revealed by neutron diffraction. Proc. Natl Acad. Sci. USA 86 , 7876–7879 (1989).
Koch, M. H. J. et al. Time-resolved X-ray diffraction study of structural changes associated with the photocycle of bacteriorhodopsin. EMBO J. 10, 521–526 (1991).
Subramaniam, S., Gerstein, M., Oesterhelt, D. & Henderson, R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 12, 1–8 (1993).
Luecke, H., Schobert, B., Richter, H. T., Cartailler, J. P. & Lanyi, J. K. Structural changes in bacteriorhodopsin during ion transport at 2 angstom resolution. Science 286, 255–261 (1999).
Lanyi, J. K. Understanding structure and function in the light-driven proton pump bacteriorhodopsin. J. Struct. Biol. 124, 164– 178 (1998).
Oesterhelt, D. The structure and mechanism of the family of retinal proteins from halophilic archaea. Curr. Opin. Struct. Biol. 8, 489 –500 (1998).
Butt, H. J., Fendler, K., Bamberg, E., Tittor, J. & Oesterhelt, D. Aspartic acids 96 and 85 play a central role in the function of bacteriorhodopsin as a proton pump. EMBO J. 8, 1657–1663 (1989).
Gerwert, K., Hess, B., Soppa, J. & Oesterhelt, D. Role of aspartate-96 in proton translocation by bacteriorhodopsin. Proc. Natl Acad. Sci. USA 86, 4943–4947 ( 1989).
Luecke, H., Schobert, B., Richter, H. T., Cartailler, J. P. & Lanyi, J. K. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 291 899–911 (1999).
Edman, K. et al. High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle. Nature 401, 822–826 (1999).
Maeda, A., Sasaki, J., Yamazaki, Y., Needleman, R. & Lanyi, J. K. Interaction of aspartate-85 with a water molecule and the protonated Schiff base in the L intermediate of bacteriorhodopsin: a Fourier-transform infrared spectroscopic study. Biochemistry 33, 1713–1717 ( 1994).
Scharnagl, C. & Fischer, S. F. Conformational flexibility of arginine-82 as source for the heterogeneous and pH-dependent kinetics of the primary proton transfer step in the bacteriorhodopsin photocycle: An electrostatic model. Chem. Phys. 212, 231– 246 (1996).
Dioumaev, A. K. et al. Existence of a proton transfer chain in bacteriorhodopsin: participation of Glu-194 in the release of protons to the extracellular surface. Biochemistry 37, 2496– 2506 (1998).
Rammelsberg, R., Huhn, G., Lübben, M. & Gerwert, K. Bacteriorhodopsin's intramolecular proton-release pathway consists of a hydrogen-bonded network. Biochemistry 37, 5001– 5009 (1998).
Zscherp, C., Schlesinger, R., Tittor, J., Oesterhelt, D. & Heberle, J. In situ determination of transient pKa changes of internal amino acids of bacteriorhodopsin by using time-resolved attenuated total reflection Fourier-transform infrared spectroscopy. Proc. Natl Acad. Sci. USA 96, 5498– 5503 (1999).
Richter, H. T., Brown, L. S., Needleman, R. & Lanyi, J. K. A linkage of the pKa's of asp-85 and glu-204 forms part of the reprotonation switch of bacteriorhodopsin. Biochemistry 35, 4054–4062 (1996).
Sass, H. J. et al. The tertiary structural changes in bacteriorhodopsin occur between M states: X-ray diffraction and Fourier transform infrared spectroscopy. EMBO J. 16, 1484–1491 (1997).
Sass, H. J. et al. Evidence for charge-controlled conformational changes in the photocycle of bacteriorhodopsin. Biophys. J. 75, 399–405 (1998).
Xu, D., Sheves, M. & Schulten, K. Molecular dynamics study of the M412 intermediate of bacteriorhodopsin. Biophys. J. 69, 2745– 2760 (1995).
Yamazaki, Y., Kandori, H., Needleman, R., Lanyi, J. K. & Maeda, A. Interaction of the protonated Schiff base with the peptide backbone of valine 49 and the intervening water molecule in the N photointermediate of bacteriorhodopsin. Biochemistry 37, 1559–1564 (1998).
Weik, M., Zaccai, G., Dencher, N. A., Oesterhelt, D. & Hauss, T. Structure and hydration of the M-state of the bacteriorhodopsin mutant D96N studied by neutron diffraction. J. Mol. Biol. 275, 625–634 (1998).
Landau, E. M. & Rosenbusch, J. P. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl Acad. Sci. USA 93, 14532–14535 (1996).
Zimanyi, L. & Lanyi, J. K. Deriving the intermediate spectra and photocycle kinetics from time- resolved difference spectra of bacteriorhodopsin. The simpler case of the recombinant D96N protein. Biophys. J. 64, 240–251 (1993).
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Luecke, H., Richter, H. T. & Lanyi, J. K. Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution. Science 280, 1934 –1937 (1998).
Genick, U. K. et al. Structure of a protein photocycle intermediate by millisecond time- resolved crystallography. Science 275, 1471–1475 (1997).
Bhat, T. N. & Cohen, G. H. OMITMAP: An electron density map suitable for the examination of errors in a macromolecular model. J. Appl. Crystallogr. 17, 244–248 (1984).
Evans, S. V. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 11, 134– 138 (1993).
Acknowledgements
We thank E. Landau, J. Rosenbusch, J. Heberle, V. Gordeliy, J. Granzin and J. Labahn for support and discussions, and A. Cousin for preparation of purple membranes. This work was supported by EU-BIOTECH and the Deutsche Forschungsgemeinschaft, Sfb 189 (H.J.S. and G.B.), the US Department of Energy, Office of Health and Enviromental Research (J.B.), and OTKA (P.O.).
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Rights and permissions
About this article
Cite this article
Sass, H., Büldt, G., Gessenich, R. et al. Structural alterations for proton translocation in the M state of wild-type bacteriorhodopsin. Nature 406, 649–653 (2000). https://doi.org/10.1038/35020607
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35020607
This article is cited by
-
True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins
Nature Structural & Molecular Biology (2022)
-
DEER Spectroscopy of Channelrhodopsin-2 Helix B Movements in Trapped Photocycle Intermediates
Applied Magnetic Resonance (2022)
-
Structure-based insights into evolution of rhodopsins
Communications Biology (2021)
-
Collective exchange processes reveal an active site proton cage in bacteriorhodopsin
Communications Biology (2020)
-
Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography
Nature Communications (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.