Abstract
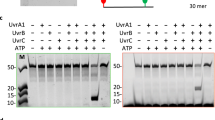
The expression of the Escherichia coli DNA polymerases pol V (UmuD′2C complex)1,2 and pol IV (DinB)3 increases in response to DNA damage4. The induction of pol V is accompanied by a substantial increase in mutations targeted at DNA template lesions in a process called SOS-induced error-prone repair4. Here we show that the common DNA template lesions, TT (6–4) photoproducts, TT cis–syn photodimers and abasic sites, are efficiently bypassed within 30 seconds by pol V in the presence of activated RecA protein (RecA*), single-stranded binding protein (SSB) and pol III's processivity β,γ-complex. There is no detectable bypass by either pol IV or pol III on this time scale. A mutagenic ‘signature’ for pol V is its incorporation of guanine opposite the 3′-thymine of a TT (6–4) photoproduct, in agreement with mutational spectra. In contrast, pol III and pol IV incorporate adenine almost exclusively. When copying undamaged DNA, pol V exhibits low fidelity with error rates of around 10-3 to 10-4, with pol IV being 5- to 10-fold more accurate. The effects of RecA protein on pol V, and β,γ-complex on pol IV, cause a 15,000- and 3,000-fold increase in DNA synthesis efficiency, respectively. However, both polymerases exhibit low processivity, adding 6 to 8 nucleotides before dissociating. Lesion bypass by pol V does not require β,γ-complex in the presence of non-hydrolysable ATPγS, indicating that an intact RecA filament may be required for translesion synthesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Tang,M. et al. Biochemical basis of SOS mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD 2′C mutagenic complex and RecA protein. Proc. Natl Acad. Sci. USA 95, 9755–9760 (1998).
Tang,M. et al. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl Acad. Sci. USA 95, 8919–8924 (1999).
Wagner,J. et al. The dinB gene encodes a novel Escherichia coli DNA polymerase (DNA pol IV). Mol. Cell 40, 281–286 (1999).
Friedberg,E. C., Walker,G. C. & Siede, W. DNA Repair and Mutagenesis 407– 522 (ASM, Washington, 1995).
Kato,T. & Shinoura,Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutagenesis by ultraviolet light. Mol. Gen. Genet. 156, 121–131 (1977).
Bonner,C. A., Hays,S., McEntee,K. & Goodman,M. F. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc. Natl Acad. Sci. USA 87, 7663–7667 (1990).
Brotocorne-Lannoye,A. & Maenhaut-Michel,G. Role of RecA protein in untargeted UV mutagenesis of bacteriophage λ: Evidence for the requirement for the dinB gene. Proc. Natl Acad. Sci. USA 83, 3904–3908 ( 1986).
Kim,S.-R. et al. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: An overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl Acad. Sci. USA 94, 13792– 13797 (1997).
Knippers,R. DNA polymerase II. Nature 228, 1050– 1053 (1970).
Rangarajan,S., Woodgate,R. & Goodman, M. F. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli . Proc. Natl Acad. Sci. USA 96, 9224 –9229 (1999).
Lindahl,T. DNA repair enzymes. Annu. Rev. Biochem. 51, 61–87 (1982).
Banerjee,S. K., Christensen,R. B., Lawrence, C. W. & LeClerc,J. E. Frequency and spectrum of mutations produced by a single cis–syn thymine–thymine cyclobutane dimer in a single-stranded vector. Proc. Natl Acad. Sci. USA 85, 8141– 8145 (1988).
Lawrence,C. W., Borden,A., Banerjee,S. K. & LeClerc,J. E. Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res. 18, 2153– 2157 (1990).
LeClerc,J. E., Borden,A. & Lawrence, C. W. The thymine–thymine pyrimidine–pyrimidone(6–4) ultravioletlight photoproduct is highly mutagenic and specifically induces 3′ thymine-to-cytosine transitions in Escherichia coli. Proc. Natl Acad. Sci. USA 88, 9685– 9689 (1991).
Rajagopalan,M. et al. Activity of the purified mutagenesis proteins UmuC, UmuD′, and RecA in replicative bypass of an abasic DNA lesions by DNA polymerase III. Proc. Natl Acad. Sci. USA 89, 10777 –10781 (1992).
Creighton,S., Bloom,L. B. & Goodman, M. F. in Methods Enzymol. Vol. 262 (ed. Campbell, J. L.) 232–256 (Academic, San Diego, 1995).
Smith,C. A. et al. Mutation spectra of M13 vectors containing site-specific cis–syn, trans–syn-I, (6–4), and dewar pyrimidone photoproducts of thymidylyl-(3′→5′)-thymidine in Escherichia coli under SOS conditions. Biochemistry 35, 4146–4154 (1996).
Lawrence,C. W., Banerjee,S. K., Borden,A. & LeClerc,J. E. T–T Cyclobutane dimers are misinstructive, rather than non-instructive, mutagenic lesions. Mol. Gen. Genet. 166, 166–168 (1990).
Fijalkowska,I. J., Dunn,R. L. & Schaaper, R. M. Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J. Bacteriol. 179, 7435–7445 ( 1997).
Bloom,L. B. et al. Fidelity of Escherichia coli DNA polymerase III holoenzyme: The effects of β, γ complex processivity proteins and ε proofreading exonuclease on nucleotide misincorporation efficiencies. J. Biol. Chem. 272, 27919–27930 ( 1997).
Sommer,S., Boudsocq,F., Devoret,R. & Bailone,A. Specific RecA amino acid changes affect RecA-UmuD′C interaction. Mol. Microbiol. 28, 281–291 ( 1998).
Menetski,J. P. & Kowalczykowski,S. C. Interaction of RecA protein with single-stranded DNA: Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 181, 281–295 (1985).
Reuven,N. B., Arad,G., Maor–Shoshani, A. & Livneh,Z. The mutagenic protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem. 274, 31763–31766 ( 1999).
Radman,M. Enzymes of evolutionary change. Nature 401, 866–869 (1999).
McDonald,J. P. et al. Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics 60, 20–30 (1999).
Gerlach,V. L. et al. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc. Natl Acad. Sci. USA 96, 11922–11927 (1999).
Naktinis,V., Turner,J. & O'Donnell, M. A molecular switch in a replication machine defined by an internal competition for protein rings. Cell 84, 137–145 (1996).
Jing,Y., Kao,J. F.-L. & Taylor, J.-S. Thermodynamic and base-pairing studies of matched and mismatched DNA dodecamer duplexes containing cis–syn, (6–4) and Dewar photoproducts of TT. Nucleic Acids Res. 26 , 3845–3853 (1998).
Bambara,R. A., Fay,P. J. & Mallaber, L. M. Methods of analyzing processivity. Methods Enzymol. 262, 270–280 ( 1995).
Bruck,I., Woodgate,R., McEntee,K. & Goodman,M. F. Purification of a soluble UmuD′C complex from Escherichia coli. Cooperative binding of UmuD′C to single-stranded DNA. J. Biol. Chem. 271, 10767–10774 ( 1996).
Acknowledgements
This work was supported by National Institutes of Health grants to M.F.G., M.O. and J.-S.T.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tang, M., Pham, P., Shen, X. et al. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404, 1014–1018 (2000). https://doi.org/10.1038/35010020
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35010020
This article is cited by
-
Ecological effects of stress drive bacterial evolvability under sub-inhibitory antibiotic treatments
ISME Communications (2022)
-
Comparative chloroplast genomics of the genus Taxodium
BMC Genomics (2020)
-
Purification and interactions of the MucA’ and MucB proteins constituting the DNA polymerase RI
Genes and Environment (2019)
-
Specialised DNA polymerases in Escherichia coli: roles within multiple pathways
Current Genetics (2018)
-
Isolating Escherichia coli strains for recombinant protein production
Cellular and Molecular Life Sciences (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.