Abstract
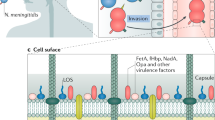
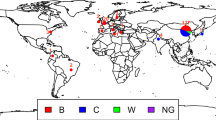
Neisseria meningitidis causes bacterial meningitis and is therefore responsible for considerable morbidity and mortality in both the developed and the developing world. Meningococci are opportunistic pathogens that colonize the nasopharynges and oropharynges of asymptomatic carriers. For reasons that are still mostly unknown, they occasionally gain access to the blood, and subsequently to the cerebrospinal fluid, to cause septicaemia and meningitis. N. meningitidis strains are divided into a number of serogroups on the basis of the immunochemistry of their capsular polysaccharides; serogroup A strains are responsible for major epidemics and pandemics of meningococcal disease, and therefore most of the morbidity and mortality associated with this disease. Here we have determined the complete genome sequence of a serogroup A strain of Neisseria meningitidis, Z2491 (ref. 1). The sequence is 2,184,406 base pairs in length, with an overall G+C content of 51.8%, and contains 2,121 predicted coding sequences. The most notable feature of the genome is the presence of many hundreds of repetitive elements, ranging from short repeats, positioned either singly or in large multiple arrays, to insertion sequences and gene duplications of one kilobase or more. Many of these repeats appear to be involved in genome fluidity and antigenic variation in this important human pathogen.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dempsey, J. A., Wallace, A. B. & Cannon, J. G. The physical map of the chromosome of a serogroup A strain of Neisseria meningitidis shows complex rearrangements relative to the chromosomes of the two mapped strains of the closely related species N. gonorrhoeae. J. Bacteriol. 177, 6390 –6400 (1995).
Curnow, A. W., Tumbula, D. L., Pelaschier, J. T., Min, B. & Soll, D. Glutamyl-tRNA (Gln) amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl Acad. Sci. USA 95, 12838– 12843 (1998).
Andersson, S. G. et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133– 140 (1998).
Lobry, J. R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 13, 660–665 (1996).
Smith, N. H., Holmes, E. C., Donovan, G. M., Carpenter, G. A. & Spratt, B. G. Networks and groups within the genus Neisseria: analysis of argF, recA, rho, and 16S rRNA sequences from human Neisseria species. Mol. Biol. Evol. 16, 773–783 (1999).
Swartley, J. S. et al. Characterization of the gene cassette required for biosynthesis of the (α1→6)-linked N-acetyl-d-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J. Bacteriol. 180, 1533–1539 ( 1998).
Goodman, S. D. & Scocca, J. J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl Acad. Sci. USA 85, 6982–6986 (1988)
Correia, F. F., Inouye, S. & Inouye, M. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem. 263, 12194–12198 ( 1988).
Haas, R. & Meyer, T. F. The repertoire of silent pilos genes in Neisseria gonorrhoeae: Evidence for gene conversion. Cell 44, 107–115 ( 1986).
Mehr, I. J., & Seifert, H. S. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30, 697–710 (1998).
Howell-Adams, B., Wainwright, L. A. & Seifert, H. S. The size and position of heterologous insertions in a silent locus differentially affect pilin recombination in Neisseria gonorrhoeae. Mol. Microbiol. 22, 509 –522 (1996).
Wainwright, L. A., Pritchard, K. H. & Seifert, H. S. A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol. Microbiol. 13, 75–87 (1994).
Pettersson, A., Prinz, T., Umar, A., van der Biezen, J. & Tommassen, J. Molecular characterization of LbpB, the second lactoferrin-binding protein of Neisseria meningitidis. Mol. Microbiol. 27, 599–610 (1998).
Vonder Haar, R. A., Legrain, M., Kolbe, H. V. & Jacobs, E. Characterization of a highly structured domain in Tbp2 from Neisseria meningitidis involved in binding to human transferrin. J. Bacteriol. 176, 6207–6213 (1994).
Lewis, L. A., Gray, E., Wang, Y. P., Roe, B. A., & Dyer, D. W. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol. Microbiol. 23, 737–749 (1997).
Adhikari, P., Berish, S. A., Nowalk, A. J., Veraldi, K. L., Morse, S. A. & Mietzner, T. A. The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J. Bacteriol. 178, 2145–2149 (1996).
Stojiljkovic, I. et. al. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15, 531–541 (1995).
Klee, S. R. et al. Molecular and biological analysis of eight genetic islands that distinguish Neisseria meningitidis from the closely related pathogen Neisseria gonorrhoeae. Infect. Immunity (in the press).
Domenighini, M. et al. Genetic characterization of Bordetella pertussis filamentous haemagglutinin: a protein processed from an unusually large precursor. Mol. Microbiol. 4, 787–800 (1990).
Henderson, I. R., Owen, P. & Nataro, J. P. Molecular switches–the ON and OFF of bacterial phase variation. Mol. Microbiol. 33, 919 –932 (1999).
Sarkari, J., Pandit, N., Moxon, E. R. & Achtman, M. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol. Microbiol. 13, 207–217 ( 1994).
van der Ende, A. et al. Variable expression of class 1 outer membrane protein in Neisseria meningitidis is caused by variation in the spacing between the-10 and-35 regions of the promoter. J. Bacteriol. 177, 2475–2480 (1995).
Zhou, J. & Spratt, B. G. Sequence diversity within the argF, fbp and recA genes of natural isolates of Neisseria meningitidis: interspecies recombination within the argF gene. Mol. Microbiol. 6, 2135– 2146 (1992).
Bonfield, J. K., Smith, K. F. & Staden, R. A new DNA sequence assembly program. Nucleic Acids Res. 23, 4992–4999 (1995).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 ( 1990).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 ( 1997).
Frishman, D., Mironov, A., Mewes, H. W. & Gelfand, M. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 26, 2941– 2947 (1998).
Salzberg, S. L., Delcher, A. L., Kasif, S. & White, O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26, 544–548 (1998).
Bateman, A. et al. Pfam 3.1: 1,313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27, 260–262 (1999).
Higgins, D. G., Bleasby, A. J. & Fuchs, R. CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci. 8, 189– 191 (1992).
Acknowledgements
This research was funded by The Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Parkhill, J., Achtman, M., James, K. et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404, 502–506 (2000). https://doi.org/10.1038/35006655
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35006655
This article is cited by
-
Structural basis for the toxic activity of MafB2 from maf genomic island 2 (MGI-2) in N. meningitidis B16B6
Scientific Reports (2023)
-
Effect of respiratory inhibitors and quinone analogues on the aerobic electron transport system of Eikenella corrodens
Scientific Reports (2021)
-
Neisseria meningitidis: using genomics to understand diversity, evolution and pathogenesis
Nature Reviews Microbiology (2020)
-
Whole-genome sequencing and characterization of an antibiotic resistant Neisseria meningitidis B isolate from a military unit in Vietnam
Annals of Clinical Microbiology and Antimicrobials (2019)
-
Transcriptomic buffering of cryptic genetic variation contributes to meningococcal virulence
BMC Genomics (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.