Abstract
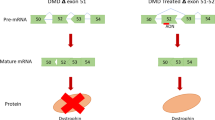
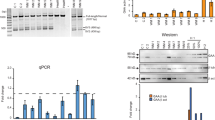
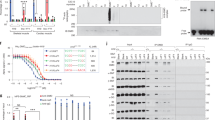
As small molecule drugs for Duchenne muscular dystrophy (DMD), antisense oligonucleotides (AONs) have been shown to restore the disrupted reading frame of DMD transcripts by inducing specific exon skipping. This allows the synthesis of largely functional Becker muscular dystrophy (BMD)-like dystrophins and potential conversion of severe DMD into milder BMD phenotypes. Thus far we have used 2′-O-methyl phosphorothioate (2OMePS) AONs. Here, we assessed the skipping efficiencies of different AON analogs containing morpholino-phosphorodiamidate, locked nucleic acid (LNA) or peptide nucleic acid (PNA) backbones. In contrast to PNAs and morpholinos, LNAs have not yet been tested as splice modulators. Compared to the most effective 2OMePS AON directed at exon 46, the LNA induced higher skipping levels in myotubes from a human control (85 versus 20%) and an exon 45 deletion DMD patient (98 versus 75%). The morpholino-induced skipping levels were only 5–6%, whereas the PNA appeared to be ineffective. Further comparative analysis of LNA and 2OMePS AONs containing up to three mismatches revealed that LNAs, while inducing higher skipping efficiencies, show much less sequence specificity. This limitation increases the risk of adverse effects elsewhere in the human genome. Awaiting further improvements in oligochemistry, we thus consider 2OMePS AONs currently the most favorable compounds, at least for targeted DMD exon 46 skipping.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Braasch DA, Corey DR . Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry 2002; 41: 4503–4510.
Suwanmanee T et al. Repair of a splicing defect in erythroid cells from patients with beta-thalassemia/HbE disorder. Mol Ther 2002; 6: 718–726.
Friedman KJ et al. Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J Biol Chem 1999; 274: 36193–36199.
Mercatante DR, Sazani P, Kole R . Modification of alternative splicing by antisense oligonucleotides as a potential chemotherapy for cancer and other diseases. Curr Cancer Drug Targets 2001; 1: 211–230.
Cartegni L, Krainer AR . Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol 2003; 10: 120–125.
Hoffman EP, Brown Jr RH, Kunkel LM . Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 1987; 51: 919–928.
Hoffman EP et al. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne's or Becker's muscular dystrophy. N Engl J Med 1988; 318: 1363–1368.
Koenig M et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet 1989; 45: 498–506.
Ervasti JM et al. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature 1990; 345: 315–319.
Di Blasi C et al. Dystrophin-associated protein abnormalities in dystrophin-deficient muscle fibers from symptomatic and asymptomatic Duchenne/Becker muscular dystrophy carriers. Acta Neuropathol (Berl) 1996; 92: 369–377.
Koenig M, Monaco AP, Kunkel LM . The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell 1988; 53: 219–226.
Dunckley MG et al. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum Mol Genet 1998; 7: 1083–1090.
Wilton SD et al. Specific removal of the nonsense mutation from the mdx dystrophin mRNA using antisense oligonucleotides. Neuromuscular Disord 1999; 9: 330–338.
De Angelis FG et al. Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Delta 48–50 DMD cells. Proc Natl Acad Sci USA 2002; 99: 9456–9461.
Takeshima Y et al. Oligonucleotides against a splicing enhancer sequence led to dystrophin production in muscle cells from a Duchenne muscular dystrophy patient. Brain Dev 2001; 23: 788–790.
van Deutekom JC et al. Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum Mol Genet 2001; 10: 1547–1554.
Aartsma-Rus A et al. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet 2003; 12: 907–914.
Aartsma-Rus A et al. Antisense-induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am J Hum Genet 2004; 74: 83–92; Epub 2003 Dec 2016.
Lu QL et al. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med 2003; 6: 6.
Agrawal S . Importance of nucleotide sequence and chemical modifications of antisense oligonucleotides. Biochim Biophys Acta 1999; 1489: 53–68.
Manoharan M . Oligonucleotide conjugates as potential antisense drugs with improved uptake, biodistribution, targeted delivery, and mechanism of action. Antisense Nucleic Acid Drug Dev 2002; 12: 103–128.
Ekker SC, Larson JD . Morphant technology in model developmental systems. Genesis 2001; 30: 89–93.
Summerton J . Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta 1999; 1489: 141–158.
Morcos PA . Achieving efficient delivery of morpholino oligos in cultured cells. Genesis 2001; 30: 94–102.
Schmajuk G, Sierakowska H, Kole R . Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J Biol Chem 1999; 274: 21783–21789.
Nasevicius A, Ekker SC . Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet 2000; 26: 216–220.
Sazani P et al. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat Biotechnol 2002; 20: 1228–1233.
Gebski BL et al. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum Mol Genet 2003; 12: 1801–1811.
Braasch DA, Liu Y, Corey DR . Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res 2002; 30: 5160–5167.
Leumann CJ . DNA analogues: from supramolecular principles to biological properties. Bioorg Med Chem 2002; 10: 841–854.
Fluiter K et al. In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res 2003; 31: 953–962.
Larsen HJ, Bentin T, Nielsen PE . Antisense properties of peptide nucleic acid. Biochim Biophys Acta 1999; 1489: 159–166.
Mologni L et al. Additive antisense effects of different PNAs on the in vitro translation of the PML/RARalpha gene. Nucleic Acids Res 1998; 26: 1934–1938.
Ray A, Norden B . Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J 2000; 14: 1041–1060.
Sazani P et al. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res 2001; 29: 3965–3974.
Karras JG et al. Deletion of individual exons and induction of soluble murine interleukin-5 receptor-alpha chain expression through antisense oligonucleotide-mediated redirection of pre-mRNA splicing. Mol Pharmacol 2000; 58: 380–387.
Aartsma-Rus A et al. Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscular Disord 2002; 12: S71.
Arzumanov A et al. Inhibition of HIV-1 Tat-dependent trans activation by steric block chimeric 2′-O-methyl/LNA oligoribonucleotides. Biochemistry 2001; 40: 14645–14654.
Acknowledgements
This work was financially supported by the Duchenne Parent Project (The Netherlands), the Princess Beatrix Fund (The Netherlands) and the MDA (USA).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Aartsma-Rus, A., Kaman, W., Bremmer-Bout, M. et al. Comparative analysis of antisense oligonucleotide analogs for targeted DMD exon 46 skipping in muscle cells. Gene Ther 11, 1391–1398 (2004). https://doi.org/10.1038/sj.gt.3302313
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.gt.3302313
Keywords
This article is cited by
-
Systematic evaluation of 2′-Fluoro modified chimeric antisense oligonucleotide-mediated exon skipping in vitro
Scientific Reports (2019)
-
Antisense oligonucleotides: the next frontier for treatment of neurological disorders
Nature Reviews Neurology (2018)
-
Antisense Oligonucleotide Mediated Splice Correction of a Deep Intronic Mutation in OPA1
Molecular Therapy - Nucleic Acids (2016)
-
Peptide Nucleic Acid Promotes Systemic Dystrophin Expression and Functional Rescue in Dystrophin-deficient mdx Mice
Molecular Therapy - Nucleic Acids (2015)
-
A Sensitive Assay System To Test Antisense Oligonucleotides for Splice Suppression Therapy in the Mouse Liver
Molecular Therapy - Nucleic Acids (2014)