Abstract
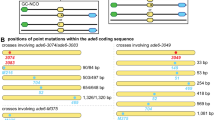
Two different models have been proposed to explain the relative frequencies of the non-mendelian allelic segregations which are detected by tetrad analysis after meiosis in fungi1–7. The first model maintains that 6:2 type tetrads result from correction of heteroduplexes containing mismatched sites and 5:3 type tetrads result from failure to correct mismatched sites. The second model suggests that 6:2 segregations result from the filling-in of double-strand gaps using information obtained from both strands of a homologous duplex. In this model 5:3 type tetrads result if the allele is included in the heteroduplex regions flanking the gap and the resulting mismatched nucleotides are not corrected7. We have studied the correction of heteroduplex plasmid DNA in pms1 mutant strains of Saccharomyces cerevisiae, which are known to exhibit higher frequencies of 5:3 type tetrads and lower frequencies of 6:2 tetrads than wild-type strains. Our results suggest that the pms1 mutation causes a defect in mismatch correction, supporting the hypothesis that meiotic gene conversion in wild-type yeast cells often results from the correction of heteroduplex DNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
1. Fogel, S., Mortimer, R. K., Lusnak, K. & Tavares, V. Cold Spring Harb. Symp. quant. Biol 43, 1325–1341 (1979). 2. Fogel, S., Mortimer, R. K. & Lusnak, K. in The Molecular Biology of the Yeast Saccharomyces Vol. 1 (eds Strathern, J. N., Jones, E. W. & Broach, J. R.) 289–339 (Cold Spring Harbor Laboratory, New York, 1982). 3. Fogel, S., Mortimer, R. K. & Lusnak, K. in Molecular and Experimental Perspectives (eds Spender, J. F. T., Spencer, D. M. & Smith, A. R. W.) 65–107 (Springer, New York, 1983). 4. Radding, C. M. A Rev. Biochem. 47, 847–880 (1978). 5. Golin, J. E. & Esposito, M. S. Molec. gen. Genet. 183, 252–263 (1981). 6. Meselson, M. S. & Radding, C. M. Proc. natn. Acad. Sci. U.S.A. 72, 358–361 (1975). 7. Szostak, J. W., Orr–Weaver, T. L., Rothstein, R. J. & Stahl, F. W. Cell 33, 25–35 (1983). 8. Williamson, M. S., Game, J. C. & Fogel, S. Genetics 110, 609–646 (1985). 9. Bishop, D. B. & Kolodner, R. D. Molec. cell. Biol. 6, 3401–3409 (1986). 10. Wagner, R. & Meselson, M. Proc. natn. Acad. Sci. U.S.A. 73, 4135–4139 (1976). 11. Stinchcomb, D. T., Mann, C. & Davis, R. W. J. molec. Biol. 158, 157–179 (1982). 12. Fishel, R. A. & Kolodner, R. /. molec. Biol. 188, 147–157 (1986). 13. Fleiss, J. L. Statistical Methods for Rates and Proportions (Wiley, New York, 1981).
Author information
Authors and Affiliations
Author notes
To whom correspondence should be addressed.
- Richard D. Kolodner
Rights and permissions
About this article
Cite this article
Bishop, D., Williamson, M., Fogel, S. et al. The role of heteroduplex correction in gene conversion in Saccharomyces cerevisiae. Nature 328, 362–364 (1987). https://doi.org/10.1038/328362a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/328362a0
This article is cited by
-
Mismatch repair genes of eukaryotes
Journal of Genetics (1996)
-
Recombinators, recombinases and recombination genes of yeasts
Current Genetics (1994)
-
Recombinational repair of diverged DNAs : a study of homoeologous chromosomes and mammalian YACs in yeast
Molecular and General Genetics MGG (1992)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.