Abstract
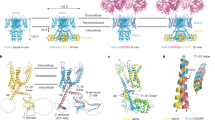
Voltage-dependent, ion-selective channels such as Na+, Ca2+ and K+ channel proteins function as tetrameric assemblies of identical or similar subunits1,2,3,4. The clustering of four subunits is thought to create an aqueous pore5,6 centred at the four-fold symmetry axis. The highly conserved, amino-terminal cytoplasmic domain (∼130 amino acids) immediately preceding the first putative transmembrane helix S1 is designated T1. It is known to confer specificity for tetramer formation7,8, so the heteromeric assembly of K+-channel subunits is an important mechanism for the observed channel diversity9,10,11. We have determined the crystal structure of the T1 domain of a Shaker potassium channel at 1.55 Å resolution. The structure reveals that four identical subunits are arranged in a four-fold symmetry surrounding a centrally located pore about 20 Å in length. Subfamily-specific assembly is provided primarily by polar interactions encoded in a conserved set of amino acids at its tetramerization interface. Most highly conserved amino acids in the T1 domain of all known potassium channels are found in the core of the protein, indicating a common structural framework for the tetramer assembly.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Noda, M. et al. Primary structure of Electrophorus electricus sodium channel deduced from cDNA sequence. Nature 312, 121–127 (1984).
Tanabe, T. et al. Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature 328, 313– 318 (1987).
Papazian, D. M., Schwarz, T. L., Tempel, B. L., Jan, Y. N. & Jan, Y. L. Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene form Drosophila. Science 237, 749–753 ( 1987).
MacKinnon, R. Determination of the subunit stoichiometry of a voltage-activated potassium channel. Nature 350, 232– 235 (1991).
Li, M., Unwin, N., Stauffer, K. A., Jan, L. Y. & Jan, Y. N. Images of purified Shaker potassium channels. Curr. Biol. 4, 110– 115 (1994).
Miller, C. 1990: Annus mirabilis of potassium channels. Science 252, 1092–1095 (1991).
Shen, N. V., Chen, X., Boyer, M. M. & Pfaffinger, P. J. Deletion analysis of K+ channel assembly. Neuron 11, 67–76 (1993).
Li, M., Jan, Y. N. & Jan, L. Y. Specification of subunit assembly by the hydrophilic amino-terminal domain of the Shaker potassium channel. Science 257, 1225–1230 (1992).
Schwarz, T. L., Tempel, B. L., Papazian, D. M., Jan, Y. N. & Jan, L. Y. Multiple potassium-channel components are produced by alternative splicing at the Shaker locus in Drosophila . Nature 331, 137–142 (1988).
Wei, A. et al. K+ current diversity is produced by an extended gene family conserved in Drosophila and mouse. Science 248, 599–603 (1990).
Stühmer, W. et al. Molecular basis of functional diversity of voltage-gated potassium channels in mammalian brain. EMBO J. 8, 3235–3244 (1989).
Holm, L. & Sander, C. Protein structure comparison by alignments of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Murzin, A. G., Brenner, S. E., Hubbard, S. T. & Chothia, C. “SCOP”: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540 (1995).
Hol, W. G., van Duijnen, P. T. & Berendsen, H. J. The alpha-helix dipole and the properties of proteins. Nature 273, 443–446 (1978).
Hoshi, T., Zagotta, W. N. & Aldrich, R. W. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science 250, 533– 538 (1990).
MacKinnon, R., Aldrich, R. W. & Lee, A. W. Functional stoichiometry of Shaker potassium channel inactivation. Science 262, 757– 759 (1993).
Foster, C. D., Chung, S., Zagotta, W. N., Aldrich, R. W. & Levitan, I. B. Apeptide derived from the ShakerB K+ channel produces short and long blocks of reconstituted Ca2+-dependent K+ channels. Neuron 9, 229–236 ( 1992).
Toro, L., Stefani, E. & Latorre, R. Internal blockade of a Ca2+-activated K+ channel by ShakerB inactivating “ball” peptide. Neuron 9, 237–245 (1992).
Antz, C. et al. NMR structure of inactivation gates from mammalian voltage-dependent potassium channels. Nature 385, 272– 274 (1997).
Isacoff, E. Y., Jan, Y. N. & Jan, L. Y. Putative receptor for the cytoplasmic inactivation gate in the Shaker K+ channel. Nature 353 , 86–90 (1991).
Liu, T., Holmgren, M., Jurman, M. E. & Yellen, G. Gated access to the pore of a voltage-dependent K+ channel. Neuron 19, 175–184 (1997).
Mannuzzu, L. M., Moronne, M. M. & Isacoff, E. Y. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science 271, 213–216 (1996).
Otwinowski, Z. in Data Collection and Processing (eds Swayer, L., Isaac, N. & Bailey, S.) 56–62 (SERC Daresbury Laboratory, Warrington, UK, 1993).
Hendrickson, W. A. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254, 51– 58 (1991).
Collaborative Computational Project, Number4. The CCP4 Suite: Programs for Crystallography. Acta Crystallogr. D 50, 670– 673 (1994).
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjelgard, M. W. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brunger, A. T. X-PLOR Version 3.8(Dept of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT, 1996).
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 ( 1994).
Evans, S. V. SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 11, 134– 138 (1993).
Nicholls, A., Sharp, K. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struc. Funct. Genet. 11, 281–296 ( 1991).
Acknowledgements
We thank H. Bellamy at beamline 1-5 at SSRL for his help; W. Yang, J. Greenwald and R. Robinson for data collection; P. Voelker for analytical centrifuging; A. Craig for mass spectroscopy; W.Fischer and C. Park for amino-acid sequencing; and G. Louie, A. Bilwes, K. Bixby, J. Noel, F. Crick and L. Orgel for discussions and comments on the manuscript. A.K. acknowledges fellowships from the Hoffman Foundation and the American Heart Association. SSRL is funded by the Department of Energy, Office of Basic Energy Science. This work was supported by grants from the NIH (P.J.P. and S.C.) and the Council for Tobacco Research (S.C.). S.C. is a Neuroscience fellow of the E.A. & J. Klingenstein Fund. C.F.S. is a HHMI investigator.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kreusch, A., Pfaffinger, P., Stevens, C. et al. Crystal structure of the tetramerization domain of the Shaker potassium channel. Nature 392, 945–948 (1998). https://doi.org/10.1038/31978
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/31978
This article is cited by
-
Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma
Biomarker Research (2022)
-
Mutagenesis of the NaChBac sodium channel discloses a functional role for a conserved S6 asparagine
European Biophysics Journal (2017)
-
The coiled-coil domain of zebrafish TRPM7 regulates Mg·nucleotide sensitivity
Scientific Reports (2016)
-
Domain Structure and Conformational Changes in rat KV2.1 ion Channel
Journal of Neuroimmune Pharmacology (2014)
-
Ionic mechanisms in pancreatic β cell signaling
Cellular and Molecular Life Sciences (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.