Abstract
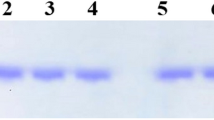
THE investigation of specific messenger RNAs in terms of sequence analysis and of the control of their synthesis, processing, translation and degradation is one of the major interests of modern molecular biology. These studies have been possible so far in only the few cases where a cell type is known to synthesise predominantly one species of protein. Since most mRNAs are chemically similar, differing only in size and sequence, it has been difficult to purify them by conventional methods. Given the amino acid sequence of a protein, one can use the genetic code to deduce a sequence of nucleotides in a specific region of the mRNA and then synthesise an oligodeoxyribonucleotide complementary to that region1–4. We have chosen to investigate the yeast iso-1 cytochrome c mRNA as a model system for the use of synthetic oligonucleotides as specific probes of eucaryotic mRNA structure and function. The genetics of yeast iso-1 cytochrome c has been studied in great detail by Sherman, Stewart and co-workers5,6, who have determined the sequence of 44 nucleotides of the mRNA from the analysis of altered cytochromes produced by a series of double frameshift mutants. In addition, a number of well defined deletion mutants are available within this region. The pentadecadeoxyribonucleotide (15-mer) that we decided to synthesise, 5′–d(A–G–C–A–C–C–T–T–T–C–T–T–A–G–C)3′ is complementary to nucleotides 25–39 of the cytochrome c mRNA (Fig. 1). This sequence was chosen because it is relatively easy to synthesise, should form a very stable hybrid with mRNA, and is close to the 5′ end of the coding region of the mRNA. A known deletion mutation covers this region, yet allows the production of a functional cytochrome c in normal amounts5,6, and thus provides an excellent control for hybridisation experiments. We have been able to show that the synthetic 15-mer does indeed bind specifically to the expected region of the mRNA, and therefore should be useful as a specific probe for the study of this mRNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Wu, R., Nature new Biol., 236, 198–200 (1972).
Wu, R., Tu, C. D., and Padmanabhan, R., Biochem. biophys. Res. Comm., 55, 1092–1099 (1973).
Narang, S. A., Itakura, K., Bahl, C. P., and Wigfield, Y. Y., Biochem. biophys. Res. Comm., 49, 445–451.
Doel, M. J., and Smith, M., FEBS Lett., 34, 99–102 (1973).
Sherman, F., and Stewart, J. N., in The Biochemistry of Gene Expression in Higher Organisms (edit. by Pollack, J. K., Lee, J. W.) 56–86 (Australian and New Zealand Book, Pty Ltd, Sydney, 1973).
Stewart, J. W., and Sherman, F., in Molecular and Environmental Aspects of Mutagenesis (edit. by Prakesh, L., Sherman, F., Miller, M. W., Lawrence, C. W., Taber, H. W.) 102–127 (Charles C. Thomas, 1974).
Itakura, K., Katagiri, N., Narang, S. A., Bahl, C. P., Marians, K. J., and Wu, R., J. biol. Chem., 250, 4592–4600 (1974).
Itakura, K., Katagiri, N., Bahl, G. P., Wightman, R. H., and Narang, S.A.,. J. Am. chem. Soc., 97, 7327–7331 (1975).
Axen, R., Porath, J., Ernback, S., Nature, 214, 1302–1304 (1967).
Katagiri, N., Itakura, K., and Narang, S. A., J. Am. chem. Soc., 97, 7332–7337 (1975).
Jay, E., Cashion, P. J., Fridkin, M., Ramamoorthy, B., Agarwal, K. M., Caruthers, M. M., and Khorana, H. G., J. biol. Chem., 251, 609–623 (1976).
Weber, H., and Khorana, H. G., J. molec. Biol., 72, 219–249 (1972).
Bahl, C. P., Wu, R., Itakura, K., Katagiri, N., and Narang, S. A., Proc. natn. Acad. Sci. U.S.A., 73, 91–94 (1976).
Stawinsky, J., Hozumi, T., and Narang, S. A., Can. J. Chem., 54, 670–672 (1976).
Jay, E., Bambara, R., Padmanabhan, R. and Wu, R., Nuc. Acids Res., 1, 331–353 (1974).
Roychoudhury, R., Jay, E., and Wu, R., Nuc. Acids Res., 3, 863–877 (1976).
Brownlee, G. G., and Sanger, F., Eur. J. Biochem., 11, 395 (1969).
Tu, C. D., Jay, E., Bahl, C. P. and Wu, R., Anal. Biochem., 74, 73–93 (1976).
Proudfoot, N. J., and Brownlee, G. G., Nature, 272, 359–362 (1974).
Gilbert, W., and Maxam, A., Proc. natn. Acad. Sci., U.S.A., in press (1977).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
SZOSTAK, J., STILES, J., BAHL, C. et al. Specific binding of a synthetic oligodeoxyribonucleotide to yeast cytochrome c mRNA. Nature 265, 61–63 (1977). https://doi.org/10.1038/265061a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/265061a0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.