Abstract
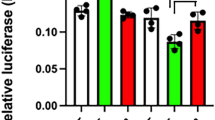
POU-domain proteins, such as the pituitary-specific factor Pit-1, are members of the homeodomain family of proteins which are important in development and homeostasis, acting constitutively or in response to signal-transduction pathways to either repress or activate the expression of specific genes1. Here we show that whereas homeodomain-containing repressors such as Rpx2 seem to recruit only a co-repressor complex, the activity of Pit-1 (ref. 3) is determined by a regulated balance between a co-repressor complex that contains N-CoR/SMRT4,5, mSin3A/B6,7,8 and histone deacetylases6,7,8 and a co-activator complex that includes the CREB-binding protein (CBP)9 and p/CAF10. Activation of Pit-1 by cyclic AMP or growth factors depends on distinct amino- and carboxy-terminal domains of CBP, respectively. Furthermore, thehistone acetyltransferase functions of CBP11,12 or p/CAF10 are required for Pit-1 function that is stimulated by cyclic AMP or growth factors, respectively. These data show that there is a switch in specific requirements for histone acetyltransferases and CBP domains in mediating the effects of different signal-transduction pathways on specific DNA-bound transcription factors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gray, S. & Levine, M. Transcriptional repression in development. Curr. Opin. Cell Biol. 8, 358–364 (1996).
Hermesz, E., Mackem, S. & Mahon, K. A. Rpx: a novel anterior-restricted homeobox gene progressively activated in the prechorda plate, anterior neural plate and Rathke's pouch of the mouse embryo. Development 122, 41–52 (1996).
Ingraham, H. A.et al. Atissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell 55, 519–529 (1988).
Horlein, A. J.et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377, 397–404 (1995).
Chen, J. D. & Evans, R. M. Atranscriptional co-repressor that interactions with nuclear hormone receptors. Nature 377, 454–457 (1995).
Heinzel, T.et al. Acomplex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387, 43–48 (1997).
Alland, L.et al. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387, 49–55 (1997).
Laherty, C. D.et al. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell 89, 349–356 (1997).
Chrivia, J. C.et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365, 855–859 (1993).
Yang, X.-Y., Ogryzko, V. V., Nishikawa, J., Howard, B. H. & Nakatani, Y. Ap300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382, 319–324 (1996).
Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. The transcriptional coactivator p300 and CBP are histone acetyltransferases. Cell 87, 953–960 (1996).
Bannister, A. J. & Kouzarides, T. The CBP coactivator is a histone acetyltransferase. Nature 384, 641–643 (1996).
Yang, W.-M., Inouye, C., Zeng, Y., Bearss, D. & Seto, E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator HDAC2/mRPD3. Proc. Natl Acad. Sci. USA 93, 12845–12850 (1996).
Kurokawa, R.et al. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature 377, 451–454 (1995).
Kamei, Y.et al. ACBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85, 403–414 (1996).
Lavinsky, R. M.et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc. Natl Acad. Sci. USA 95, 2920–2925 (1998).
Lundblad, J. R., Kwok, R. P., Laurance, M. E., Harter, M. L. & Goodman, R. H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature 374, 85–88 (1995).
Glass, C. K., Lipkin, S. M., Devary, O. V. & Rosenfeld, M. G. Positive and negative regulation of gene transcription by a retinoic acid–thyroid hormone receptor heterodimer. Cell 59, 697–708 (1989).
Jacobson, E. M., Li, P., Leon-del-Rio, A., Rosenfeld, M. G. & Aggarwal, A. K. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev. 11, 198–212 (1997).
Elsholtz, H. P., Lew, A. M., Albert, P. R. & Sundmark, V. C. Inhibitory control of prolactin and Pit-1 gene promoters by dopamine. J. Biol. Chem. 266, 22919–22925 (1991).
Murdoch, G. H., Waterman, M., Evans, R. M. & Rosenfeld, M. G. Molecular mechanism of phorbol ester, thyrotropin-releasing hormone, and growth factor stimulation of prolactin gene transcription. J. Biol. Chem. 260, 11852–11858 (1985).
Howard, P. W. & Maurer, R. A. Acomposite Ets/Pit-1 binding site in the prolactin gene can mediate transcriptional responses to multiple signal transduction pathways. J. Biol. Chem. 270, 20930–20936 (1995).
Okimura, Y., Howard, P. W. & Maurer, R. A. Pit-1 binding sites mediate transcriptional responses tocyclic adenosine 3′,5′-monophosphate through a mechanism that does not require inducible phosphorylation of Pit-1. Mol. Endocrinol. 8, 1559–1565 (1994).
Kapiloff, M. S., Farkash, Y., Wegner, M. & Rosenfeld, M. G. Variable effects of phosphorylation of Pit-1 dictated by the DNA response elements. Science 253, 786–789 (1991).
Õnate, S. A., Tsai, S. Y., Tsai, M.-J. & O'Malley, B. W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270, 1354–1357 (1995).
Torchia, J.et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387, 677–684 (1997).
Korzus, E.et al. Transcriptional factor-specific requirements for coactivators and their acetyltransferase functions. Science 279, 703–707 (1998).
Rose, D. W., McCabe, G., Feramisco, J. R. & Adler, M. Expression of c-fos and AP-1 activity in senescent human fibroblasts is not sufficient for DNA synthesis. J. Cell. Biol. 119, 1405–1411 (1992).
Acknowledgements
We thank C. Laherty, R. Eisenman, W.-M. Yang and E. Seto for antibodies, C. Nelson for cell culture, L.-M. Philips for assistance, and P. Meyer for her expertise in the preparation of the illustrations. This work was supported by an American Heart Association Predoctoral fellowship (L.X.), the NSF (R.M.L.), the Lucille Markey Fund (J.S.D.), and an NIH predoctoral fellowship (D.S.), a US Army Medical Research Program Award (E.K.), by grants from HHMI and NIH (to M.G.R.), and by an American Diabetes Association Career Development Award (to D.W.R.). C.K.G. is an Established Investigator of the American Heart Association and M.G.R. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Rights and permissions
About this article
Cite this article
Xu, L., Lavinsky, R., Dasen, J. et al. Signal-specific co-activator domain requirements for Pit-1 activation. Nature 395, 301–306 (1998). https://doi.org/10.1038/26270
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/26270
This article is cited by
-
A Pit-1 Binding Site Adjacent to E-box133 in the Rat PRL Promoter is Necessary for Pulsatile Gene Expression Activity
Neurochemical Research (2016)
-
Required enhancer–matrin-3 network interactions for a homeodomain transcription program
Nature (2014)
-
A novel recessive splicing mutation in the POU1F1 gene causing combined pituitary hormone deficiency
Journal of Endocrinological Investigation (2009)
-
Altered Memory Capacities and Response to Stress in p300/CBP-Associated Factor (PCAF) Histone Acetylase Knockout Mice
Neuropsychopharmacology (2008)
-
HDAC3: taking the SMRT-N-CoRrect road to repression
Oncogene (2007)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.