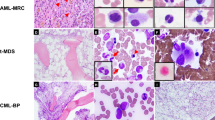
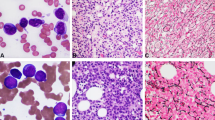
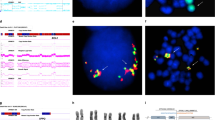
Abstract
Fluorescence in situ hybridization and comparative genomic hybridization characterized 6p rearrangements in eight primary and in 10 secondary myeloid disorders (including one patient with Fanconi anemia) and found different molecular lesions in each group. In primary disorders, 6p abnormalities, isolated in six patients, were highly heterogeneous with different breakpoints along the 6p arm. Reciprocal translocations were found in seven. In the 10 patients with secondary acute myeloid leukemia/myelodysplastic syndrome (AML/MDS), the short arm of chromosome 6 was involved in unbalanced translocations in 7. The other three patients showed full or partial trisomy of the 6p arm, that is, i(6)(p10) (one patient) and dup(6)(p) (two patients). In 5/7 patients with unbalanced translocations, DNA sequences were overrepresented at band 6p21 as either cryptic duplications (three patients) or cryptic low-copy gains (two patients). In the eight patients with cytogenetic or cryptic 6p gains, we identified a common overrepresented region extending for 5–6 megabases from the TNF gene to the ETV-7 gene. 6p abnormalities were isolated karyotype changes in four patients. Consequently, in secondary AML/MDS, we hypothesize that 6p gains are major pathogenetic events arising from acquired and/or congenital genomic instability.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Soekarman D, von Lindern M, Daenen S, de Jong B, Fonatch C, Heinze B et al. The translocation (6;9)(p23;q34) shows consistent rearrangement of two genes and defines a myeloproliferative disorder with specific clinical features. Blood 1992; 79: 2990–2997.
Oyarzo MP, Lin P, Glassman A, Bueso-Ramos CE, Luthra R, Medeiros J . Acute myeloid leukemia with t(6;9)(p23;q34) is associated with dysplasia and a high frequency of FLT3 mutations. Hematopathology 2004; 122: 348–358.
Akasaka T, Miura I, Takahashi N, Akasaka H, Yonetani N, Ohno H et al. A recurring translocation, t(3;6)(q27;p21), in non-Hodgkin's lymphoma results in replacement of the 5′ regulatory region of BCL-6 with a novel H4 histone gene. Cancer Res 1997; 57: 7–12.
Chen W, Itoyama T, Chaganti RSK . Splicing factor SRP20 is a novel partner of BCL6 in a t(3;6)(q27;p21) translocation in transformed follicular lymphoma. Genes Chromosome Cancer 2001; 32: 281–284.
Sonoki T, Harder L, Horsman DE, Karran L, Taniguchi I, Willis TG et al. Cyclin D3 is a target of t(6;14)(p21.1;q32.3) of mature B-cell malignancies. Blood 2001; 98: 2837–2844.
Shaughnessy J, Gabrera A, Qi Y, Brents L, Zhan F, Tian E et al. Cyclin D3 at 6p21 is dysregulated by recurrent chromosomal translocations to immunoglobulin loci in multiple myeloma. Blood 2001; 98: 217–223.
Iida S, Rao PH, Butler M, Corradini P, Boccadoro M, Klein B et al. Deregulation of MUM/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet 1997; 17: 226–230.
Johansson B, Mertens F, Heim S, Kristoffersson U, Mitelman F . Cytogeentics of secondary myelodysplasia (sMDS) and acute nonlymphocytic leukemia (sANLL). Eur J Haematol 1991; 47: 17–27.
Mecucci C, Michaux J-L, Louwagie A, Boogaerts M, Van Den Berghe H . The short arm of chromosome 6 is nonrandomly rearranged in secondary myelodysplastic syndromes. Cancer Genet Cytogenet 1988; 31: 147–155.
Huret JL, Schoenwald M, Brizard A, Guilhot F, Vilmer E, Tanzer J . Chromosome 6p rearrangements appear to be secondary changes in various haematological malignancies. Leukemia Res 1989; 13: 819–824.
Mancini M, Mecucci C, Cedrone M, Rondinelli MB, Aloe-Spiriti A, Alimena G . Unbalanced 6p translocation as primary karyotipic anomaly in secondary acute nonlymphocytic leukemia. Cancer Genet Cytogenet 1992; 60: 93–95.
Jacob AK, Sreekantaiah C, Baer MR, Sandberg AA . Translocation (1;6)(p12;p23) in ANLL. Cancer Genet Cytogenet 1990; 45: 67–71.
Michalovà K, Lemez P, Bartsch O, Brezinova J, Zemanova Z, Jelinek J et al. Derivative (6)t(1;6)(q22;p21) revealed in bone marrow cells by FISH 9 months before diagnosis of acute T-lymphoblastic leukemia. Cancer Genet Cytogenet 1996; 86: 131–135.
Mathew S, Head D, Rodriguez-Galindo C, Raimondi S . Trisomy of the long arm of chromosome 1 resulting in a dicentric derivative (6)t(1;6) chromosome in a child with myelodysplastic syndrome following treatment for a primitive neuroectodermal tumor. Leukemia Lymphoma 2000; 37: 213–218.
Mitelman F (ed) An International System for Human Cytogenetic Nomenclature (ISCN). Basel: S Karger, 1995.
Crescenzi B, La Starza R, Romoli S, Beacci D, Matteucci C, Barba G et al. Submicroscopic deletions in 5q− associated malignancies. Haematologica 2004; 89: 281–285.
La Starza R, Specchia G, Cuneo A, Beacci D, Nozzoli C, Luciano L et al. The hypereosinophilic syndrome: fluorescence in situ hybridization detects the del(4)(q12)-FIP1L1/PDGFRA but not genomic rearrangements of other tyrosine kinases. Haematologica 2005; 90: 596–601.
Matteucci C, La Starza R, Crescenzi B, Falzetti D, Romoli S, Emiliani C et al. Interpretation of the complex karyotype and identification of a new 6p amplicon by integrated comparative genomic hybridization and fluorescence in situ hybridization investigations on the U937-I cell line. Cancer Genet Cytogenet 2002; 135: 28–34.
Mauritzson N, Albin M, Rylander L, Billstrom R, Ahlgren T, Mikoczy Z et al. Pooled analysis of clinical and cytogenetic features in treatment-related and de novo adult acute myeloid leukemia and myelodysplastic syndromes based on a consecutive series of 761 patients analyzed 1976–1993 and on 5098 unselected cases reported in the literature 1974–2001. Leukemia 2002; 16: 2366–2378.
Sasaki MS, Tonomura A . A high susceptibility of Fanconi's anemia to chromosome breakage by DNA cross-linking agents. Cancer Res 1973; 33: 1829–1836.
Cuthbert G, Thompson K, McCullough S, Watmore A, Dickinson H, Telford N et al. MLL amplification in acute leukaemia: a United Kingdom Cancer Cytogenetics Group (UKCCG) study. Leukemia 2000; 14: 1885–1891.
Mathew S, Lorsbach RB, Shearer P, Sandlund JT, Raimondi SC . Double minute chromosomes and c-MYC amplification in a child with secondary myelodysplastic syndrome after treatment for acute lymphoblastic leukemia. Leukemia 2000; 14: 1314–1315.
Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S et al. Recent segmental duplications in the human genome. Science 2002; 297: 1003–1007.
Stankiewicz P, Lupski JR . Genome architecture, rearrangements and genomic disorders. Trends Genet 2002; 18: 74–82.
Ji Y, Eicher EE, Schwartz S, Nicholls RD . Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res 2000; 10: 597–610.
Saglio G, Storlazzi CT, Giugliano E, Surace C, Anelli L, Rege-Cambrin G et al. A 76 Kb duplicon maps close to the BCR gene on chromosome 22 and the ABL gene on chromosome 9: possible involvement in the genesis of the philadelphia chromosome translocation. Proc Natl Acad Sci USA 2002; 99: 9882–9887.
Giglio S, Broman KW, Matsumoto N, Calvari V, Gimelli G, Neumann T et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 2001; 68: 874–883.
Giglio S, Calvari V, gregato G, Gimelli G, Camanini S, Giorda R et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet 2002; 71: 276–285.
Rigaud G, Moore PS, Taruscio D, Scardoni M, Montresor M, Menestrina F et al. Alteration of chromosome arm 6p is characteristic of primary mediastinal B-cell lymphoma, as identified by genome-wide allelotyping. Genes Chromosomes Cancer 2001; 31: 191–195.
Lau CC, Harris CP, Lu X-Y, Perloky L, Gogineni S, Chintagumpala M et al. Frequent amplification and rearrangement of chromosomal bands 6p12p21 and 17p11.2 in osteosarcoma. Genes Chromosomes Cancer 2004; 39: 11–21.
Bjorkqvist AM, Tammilehto L, Nordling S, Nurminen M, Anttilla S, Mattson K et al. Comparison of DNA copy number changes in malignant mesothelioma, adenocarcinoma and large-cell anaplastic carcinoma of the lung. Br J Cancer 1998; 77: 260–269.
Nessling M, Richter K, Schwaenen C, Roerig P, Wrobel G, Wessendorf S et al. Candidate genes in breast cancer revealed by microarray-based comparative genomic hybridization of archived tissue. Cancer Res 2005; 65: 439–447.
Hauptmann S, Denkert C, Koch I, Petersen S, Scluns K, Reles A et al. Genetic alterations in epithelial ovarian tumors analyzed by comparative genomic hybridization. Hum Pathol 2002; 33: 632–641.
Chen D, Pajovic S, Duckett A, Brown VD, Squire JA, Gallie BL . Genomic amplification in retinoblastoma narrowed to 0.6 megabase on chromosome 6p containing a kinesin-like gene, RBKIN. Cancer Res 2002; 62: 967–971.
Aalto Y, Eriksson L, Seregard S, Larsson O, Knuutila S . Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Invest Ophthalmol Vis Sci 2001; 42: 313–317.
Tchinda J, Dijkhuizen T, van der Vlies P, Kok K, Horst J . Translocations involving 6p22 in acute myeloid leukaemia at relapse: breakpoint characterization using microarray-based comparative genomic hybridization. Br J Haematol 2004; 126: 495–500.
Sornberger KS, Weremowicz S, Williams AJ, Quade BJ, Ligon AH, Pedeutour F et al. Expression of HMGIY in three uterine leiomyomata with complex rearrangements of chromosome 6. Cancer Genet Cytogenet 1999; 114: 9–16.
Giraudier S, Chagraoui H, Komura E, Barnache S, Blanchet B, LeCouedic JP et al. Overexpression of FKBP51 in idiopathic myelofibrosis regulates the growth factor independence of megakaryocyte progenitors. Blood 2002; 100: 2932–2940.
Andersen MK, Christiansen DH, Kirchhoff M, Pedersen-Bjergaard J . Duplication or amplification of chromosome 11q23, including the unrearranged MLL gene, is a recurrent abnormality in therapy-related MDS and AML, and is closely related to mutation of the TP53 gene and to previous therapy with alkylating agents. Genes Chromosomes Cancer 2001; 31: 337–349.
Andersen MK, Christiansen DH, Pedersen-Bjegaard J . Amplification or duplication of chromosome band 21q22 with multiple copies of the AML1 gene and mutation of the TP53 gene in therapy-related MDS and AML. Leukemia 2005; 19: 197–200.
Acknowledgements
PAC and BAC clones were obtained from the Roswell Park Cancer Institute libraries RPCI-1, RPCI-3, RPCI-5 and RPCI-11, http://www.chori.org/BACPAC. RP5-1106L7 was kindly provided by Dr M Rocchi, University of Bari, Italy; cosmid Cah5 by Dr E Weiss, Ludwig Maximilians Universität, München, Germany; and clones RP1-22O11, RP1-99J17, RP1-162J16, RP1-124L9 and RP3-329A5 by Dr I Ragoussis King's College, London, UK. We thank Dr Geraldine Anne Boyd for assistance in preparing the manuscript. This work was supported by AIRC (Associazione Italiana Ricerca sul Cancro), CNR (Consiglio Nazionale delle Ricerche), MIUR (Ministero per l’Istruzione, l’Università e la Ricerca Scientifica), Fondazione Cassa di Risparmio, Perugia, Associazione ‘Sergio Luciani’, Fabriano, AULL (Associazione Umbra contro le Leucemie e Linfomi) Italy; and Belgian Programme of Interuniversity Poles of Attraction initiated by Belgian State, Prime Minister's Office, Science Policy Programming. BC is supported by a grant from FIRC (Fondazione Italiana per la Ricerca sul Cancro).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
La Starza, R., Aventin, A., Matteucci, C. et al. Genomic gain at 6p21: a new cryptic molecular rearrangement in secondary myelodysplastic syndrome and acute myeloid leukemia. Leukemia 20, 958–964 (2006). https://doi.org/10.1038/sj.leu.2404208
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2404208
Keywords
This article is cited by
-
Association between acute myeloid leukemia and isochromosome 6p: a case study and review of the literature
Annals of Hematology (2010)