Abstract
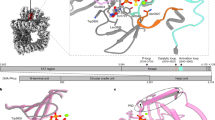
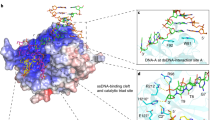
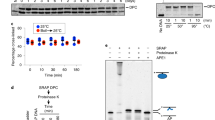
The anticancer activity of cis -diamminedichloroplatinum(II) (cisplatin) arises from its ability to damage DNA, with the major adducts formed being intrastrand d(GpG) and d(ApG) crosslinks1. These crosslinks bend and unwind the duplex, and the altered structure attracts high-mobility-group domain (HMG) and other proteins2. This binding of HMG-domain proteins to cisplatin-modified DNA has been postulated to mediate the antitumour properties of the drug3,4. Many HMG-domain proteins recognize altered DNA structures such as four-way junctions and cisplatin-modified DNA5, but until now the molecular basis for this recognition was unknown. Here we describe mutagenesis, hydroxyl-radical footprinting and X-ray studies that elucidate the structure of a 1:1 cisplatin-modified DNA/HMG-domain complex. Domain A of the structure-specific HMG-domain protein HMG1 binds to the widened minor groove of a 16-base-pair DNA duplex containing a site-specific cis -[Pt(NH3)2{d(GpG)-N7(1),-N7(2)}] adduct. The DNA is strongly kinked at a hydrophobic notch created at the platinum–DNA crosslink and protein binding extends exclusively to the 3′ side of the platinated strand. A phenylalanine residue at position 37 intercalates into a hydrophobic notch created at the platinum crosslinked d(GpG) site and binding of the domain is dramatically reduced in a mutant in which alanine is substituted for phenylalanine at this position.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lepre, C. A. & Lippard, S. J. in Nucleic Acids and Molecular Biology(eds Eckstein, F. & Lilley, D. M. J.) 9–38 (Springer, Heidelberg, 1990).
Zlatanova, J., Yaneva, J. & Leuba, S. H. Proteins that specifically recognize cisplatin-damaged DNA: aclue to anticancer activity of cisplatin. FASEB J. 12, 791–799 (1998).
Zamble, D. B. & Lippard, S. J. Cisplatin and DNA repair in cancer chemotherapy. Trends Biochem. Sci. 20, 435–439 (1995).
Zhai, X., Beckmann, H., Jantzen, H.-M. & Essigmann, J. M. Cisplatin–DNA adducts inhibit ribosomal RNA synthesis by hijacking the transcription factor human upstream binding factor. Biochemistry 37, 16307–16315 (1998).
Bustin, M. & Reeves, R. in Progr. Nucleic Acid Res. Mol. Biol.(eds Cohn, W. E. & Moldave, K.) 35–100 (Academic, San Diego, 1996).
Read, C. M. et al. . in Nucleic Acids and Molecular Biology(eds Eckstein, F. & Lilley, D. M. J.) 222–250 (Springer, Berlin, 1995).
Dunham, S. U. & Lippard, S. J. DNA sequence context and protein composition modulate HMG-domain protein recognition of cisplatin-modified DNA. Biochemistry 36, 11428–11436 (1997).
Chow, C. S., Barnes, C. M. & Lippard, S. J. Asingle HMG domain in high-mobility group 1 protein binds to DNAs as small as 20 base pairs containing the major cisplatin adduct. Biochemistry 34, 2956–2964 (1995).
Werner, M. H., Huth, J. R., Gronenborn, A. M. & Clore, G. M. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY–DNA complex. Cell 81, 705–714 (1995).
Love, J. J. et al. . Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376, 791–795 (1995).
Lavery, R. & Sklenar, H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 6, 63–91 (1988).
Dunham, S. U., Dunham, S. U., Turner, C. J. & Lippard, S. J. Solution structure of a DNA duplex containing a nitroxide spin-labeled platinum d(GpG) intrastrand cross-link refined with NMR-derived long-range electron–proton distance restraints. J. Am. Chem. Soc. 120, 5395–5406 (1998).
Gelasco, A. & Lippard, S. J. NMR solution structure of a DNA dodecamer duplex containing a cis -diammineplatinum(II) d(GpG) intrastrand cross-link, the major adduct of the anticancer drug cisplatin. Biochemistry 37, 9230–9239 (1998).
Takahara, P. M., Frederick, C. A. & Lippard, S. J. Crystal structure of the anticancer drug cisplatin bound to duplex DNA. J. Am. Chem. Soc. 118, 12309–12321 (1996).
Hardman, C. H. et al. . Structure of the A-domain of HMG1 and its interaction with DNA as studied byheteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry 34, 16596–16607 (1995).
Broadhurst, R. W., Hardman, C. H., Thomas, J. O. & Laue, E. D. Backbone dynamics of the A-domain of HMG1 as studied by 15N NMR spectroscopy. Biochemistry 34, 16608–16617 (1995).
Sherman, S. E., Gibson, D., Wang, A. H.-J. & Lippard, S. J. Crystal and molecular structure of cis -[Pt(NH3)2{d(pGpG)}], the principal adduct formed by cis -diamminedichloroplatinum(II) with DNA. J. Am. Chem. Soc. 110, 7368–7381 (1988).
Dickerson, R. E. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 26, 1906–1926 (1998).
Yang, D., van Boom, S. S. G. E., Reedijk, J., van Boom, J. H. & Wang, A. H.-J. Structure and isomerization of an intrastrand cisplatin-corsslinked octamer DNA duplex by NMR analysis. Biochemistry 34, 12912–12920 (1995).
Teo, S.-H., Grasser, K. D. & Thomas, J. O. Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex-determining factor SRY. Eur. J. Biochem. 230, 943–950 (1995).
Weiss, M. A., Ukiyama, E. & King, C.-Y. The SRY cantilever motif discriminates between sequence- and structure-specific DNA recognition: alanine mutagenesis of an HMG box. J. Biomol. Struct. Dyn. 15, 177–184 (1997).
Berners-Price, S. J. et al. . Structural transitions of a GG-platinated DNA duplex induced by pH, temperature and box A of high-mobility group protein 1. Eur. J. Biochem. 243, 782–791 (1997).
Balaeff, A., Churchill, M. E. A. & Schulten, K. Structure prediction of a complex between the chromosomal protein HMG-D and DNA. Proteins 30, 113–135 (1998).
Yoshioka, K. et al. . Differences in DNA recognition and conformational change activity between boxes A and B in HMG2 protein. Biochemistry 38, 589–595 (1999).
Falciola, L., Murchie, A. I. H., Lilley, D. M. J. & Bianchi, M. E. Mutational analysis of the DNA binding domain A of chromosomal protein HMG1. Nucleic Acids Res. 22, 285–292 (1994).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Rould, M. A. Screening for heavy-atom derivatives and obtaining accurate isomorphous differences. Methods Enzymol. 276, 461–472 (1997).
Collaborative Computational Project number 4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Brünger, A. T. XPLOR Version 3.1, A system for X-ray crystallography and NMR (Yale University Press, New Haven, Connecticut, 1992).
Baxevanis, A. D. & Landsman, D. The HMG-1 box protein family: classification and functional relationships. Nucleic Acids Res. 23, 1604–1613 (1995).
Acknowledgements
This work was supported by a grant from the National Cancer Institute (to S.J.L.), by the Howard Hughes Medical Institute (C.O.P.), by equipment purchased with support from the PEW Charitable Trusts, and by predoctoral fellowships from the Fonds der Chemischen Industrie, Germany (U.-M.O.), and the Howard Hughes Medical Institute (Q.H.). We thank S. U. Dunham for discussion, and J. S. Wishnok for mass spectrometric analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ohndorf, UM., Rould, M., He, Q. et al. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature 399, 708–712 (1999). https://doi.org/10.1038/21460
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/21460
This article is cited by
-
Understanding the Interactions of High-Mobility Group of Protein Domain B1 with DNA Adducts Generated by Platinum Anticancer Molecules Using In Silico Approaches
Interdisciplinary Sciences: Computational Life Sciences (2018)
-
Conformational rearrangement of 1,2-d(GG) intrastrand cis-diammineplatinum crosslinked DNA is driven by counter-ion penetration within the minor groove of the modified site
Journal of Molecular Modeling (2017)
-
EGCG in Green Tea Induces Aggregation of HMGB1 Protein through Large Conformational Changes with Polarized Charge Redistribution
Scientific Reports (2016)
-
A platinum-based hybrid drug design approach to circumvent acquired resistance to molecular targeted tyrosine kinase inhibitors
Scientific Reports (2016)
-
Nucleolar control of p53: a cellular Achilles’ heel and a target for cancer therapy
Cellular and Molecular Life Sciences (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.