Abstract
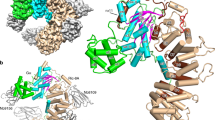
The protein Ran is a small GTP-binding protein that binds to two types of effector inside the cell: Ran-binding proteins, which have a role in terminating export processes from the nucleus to the cytoplasm, and importin-β-like molecules that bind cargo proteins during nuclear transport. The Ran-binding domain is a conserved sequence motif found in several proteins that participate in these transport processes. The Ran-binding protein RanBP2 contains four of these domains and constitutes a large part of the cytoplasmic fibrils that extend from the nuclear-pore complex. The structure of Ran bound to a non-hydrolysable GTP analogue (Ran·GppNHp) in complex with the first Ran-binding domain (RanBD1) of human RanBP2 reveals not only that RanBD1 has a pleckstrin-homology domain fold, but also that the switch-I region of Ran·GppNHp resembles the canonical Ras·GppNHp structure and that the carboxy terminus of Ran is wrapped around RanBD1, contacting a basic patch on RanBD1 through its acidic end. This molecular ‘embrace’ enables RanBDs to sequester the Ran carboxy terminus, triggering the dissociation of Ran·GTP from importin-β-related transport factors and facilitating GTP hydrolysis by the GTPase-activating protein ranGAP. Such a mechanism represents a new type of switch mechanism and regulatory protein–protein interaction for a Ras-related protein.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Görlich, D. Transport into and out of the cell nucleus. EMBO J. 17, 2721–2727 (1998).
Hartmann, E. & Görlich, D. ARan-binding motif found in nuclear pore proteins. Trends Cell Biol. 5, 192–193 (1995).
Dingwall, C., Kandels-Lewis, S. & Séraphin, B. Afamily of Ran binding proteins that includes nucleoporins. Proc. Natl Acad. Sci. USA 92, 7525–7529 (1995).
Rexach, M. & Blobel, G. Protein import into nuclei—association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83, 683–692 (1995).
Görlich, D., Pante, N., Kutay, U., Aebi, U. & Bischoff, F. R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15, 5584–5594 (1996).
Fornerod, M., Ohno, M., Yoshida, M. & Mattaj, I. W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051–1060 (1997).
Floer, M. & Blobel, G. The nucleic transport factor karyopherin-beta binds stoichiometrically to ran-GTP and inhibits the ran GTPase-activating protein. J. Biol. Chem. 271, 5313–5316 (1996).
Bischoff, F. R., Krebber, H., Smirnova, E., Dong, W. & Ponstingl, H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 14, 705–715 (1995).
Bischoff, F. R. & Görlich, D. RanBP1 is crucial for the release of Ran·GTP from importin-β-related nuclear transport factors. FEBS lett. 419, 249–254 (1997).
Wilken, N., Senécal, J.-L., Scheer, U. & Dabauvalle, M.-C. Localization of the Ran-GTP binding protein RanBP2 at the cytoplasmic side of the nuclear pore complex. Eur. J. Cell Biol. 68, 211–219 (1995).
Delphin, C., Guan, T., Melchior, F. & Gerace, L. Ran·GTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol. Biol. Cell 8, 2379–2390 (1997).
Yokoyama, N.et al. Agiant nucleopore protein that binds Ran/TC4. Nature 376, 184–188 (1995).
Wu, J., Matunis, M. J., Kraemer, D., Blobel, G. & Coutavas, E. Nup358, a cytoplasmatically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270, 14209–14213 (1995).
Radu, A., Moore, M. S. & Blobel, G. The peptide repeat domain of nucleoporin Nup98 functions as docking site in transport across the nuclear pore complex. Cell 81, 215–222 (1995).
Melchior, F., Guan, T., Yokoyama, N., Nishimoto, T. & Gerace, L. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J. Cell Biol. 131, 571–581 (1995).
Melchior, F. & Gerace, L. Two-way trafficking with Ran. Trends Cell Biol. 8, 175–179 (1998).
Scheffzek, K., Klebe, C., Fritz-Wolf, K., Kabsch, W. & Wittinghofer, A. Crystal structure of human Ran complexed with GDP. Nature 374, 378–381 (1995).
Amor, J. C.et al. Structure of the human ADP-ribosylation factor I complexed with GDP. Nature 372, 704–708 (1994).
Greasley, S. E.et al. The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms. Nature Struct. Biol. 2, 797–806 (1995).
Abel, K., Yoder, M. D., Hilgenfeld, R. & Jurnak, F. An alpha to beta conformational switch in EF-TU. Structure 4, 1153–1159 (1996).
Polekhina, G.et al. Helix unwinding in the effector of elongation factor EF-TU-GDP. Structure 4, 1141–1151 (1996).
Wittinghofer, A. & Pai, E. F. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem. Sci. 16, 382–387 (1991).
Goldberg, J. Structural basis for activation of ARF GTPase: Mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell 95, 237–248 (1998).
Richards, S. A., Lounsbury, K. M. & Macara, I. The C terminus of the nuclear RAN/TC4 GTPase stabilizes the GDP-bound state and mediates interactions with RCC1, RAN-GAP, and HTF9A/RANBP1. J. Biol. Chem. 270, 14405–14411 (1995).
Klebe, C., Bischoff, F. R., Ponstingl, H. & Wittinghofer, A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry 34, 639–647 (1995).
Ribbeck, K., Lipowsky, G., Kent, H. M., Stewart, M. & Görlich, D. NTF2 mediates nuclear import of Ran. EMBO J. 17, 6587–6598 (1998).
Stewart, M., Kent, H. M. & Mccoy, A. J. Structural basis for molecular recognition between nuclear transport factor 2 (NFT2) and GTP-bound form of the Ras family GTPase Ran. J. Mol. Biol. 277, 535–646 (1998).
Beddow, A. L., Richards, S. A., Orem, N. R. & Macara, I. G. The Ran/TC4 GTPase-binding domain: Identification by expression cloning and characterization of a conserved sequence motif. Proc. Natl Acad. Sci. USA 92, 3328–3332 (1995).
Saraste, M. & Hyvönen, M. Pleckstrin homology domains: a fact file. Curr. Opin. Struct. Biol. 5, 403–408 (1995).
Hyvönen, M. & Saraste, M. Structure of the PH domain and BTK motif from Bruton's tyrosine kinase: Molecular explanations for X-linked agammaglobulinaemia. EMBO J. 16, 3396–3404 (1997).
Touhara, K., Inglese, J., Pitcher, J. A., Shaw, G. & Lefkowitz, R. J. Binding of G protein βγ-subunits to pleckstrin homology domains. J. Biol. Chem. 269, 10217–10220 (1994).
Schlenstedt, G., Wong, D. H., Koepp, D. M. & Silver, P. A. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 14, 5367–5378 (1995).
Connolly, M. L. Analytical molecular surface calculation. J. Appl. Crystallogr. 16, 548–558 (1983).
Kuhlmann, J., Macara, I. & Wittinghofer, A. Dynamic and equilibrium studies on the interaction of Ran with its effector RanBP1. Biochemistry 36, 12027–12035 (1997).
Ferguson, K. M., Lemmon, M. A., Schlessinger, J. & Sigler, P. B. Structure of the high affinity complex of inositol triphosphate with a phospholipase C pleckstrin homology domain. Cell 83, 1037–1046 (1995).
Hyvönen, M.et al. Structure of the binding site for inositol phosphates in a PH domain. EMBO J. 14, 4676–4685 (1995).
Herrmann, C., Martin, G. A. & Wittinghofer, A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J. Biol. Chem. 270, 2901–2905 (1995).
Nassar, N.et al. The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature 375, 554–560 (1995).
Lounsbury, K. M., Richards, S. A., Carey, K. L. & Macara, I. G. Mutations within the Ran/TC4 GTPase-effects on regulatory factor interactions and subcellular localization. J. Biol. Chem. 271, 32834–32841 (1996).
Scheffzek, K.et al. The Ras–RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 (1998).
Chi, N. C., Adam, E. J. H., Visser, G. D. & Adam, S. A. RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J. Cell. Biol. 135, 559–569 (1996).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell contents. J. Appl. Crystallogr. 26, 795–800 (1993).
Collaborative Computational Project No. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Jones, T. A. & Kjeldgaard, M. Electron-density map interpretation. Methods Enzymol. 277, 173–208 (1997).
Brunger, A. T.et al. Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr. C 54, 905–921 (1998).
Esnouf, R. M. An extensively modified version of MOLSCRIPT that includes greatly enhanced coloring capabilities. J. Mol. Graphics 15, 132–134 (1997).
Pai, E. F.et al. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 9, 2351–2359 (1990).
Kraulis, P. J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E. A. & Murphy, M. E. P. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994).
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991).
Acknowledgements
We thank A. Scherer for help with the original crystallization, R. Schebaum for secretarial assistance, and the staff of the EMBL outstation in Hamburg for help with the data collection. This study was supported by HFSP and the EG (A.W.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vetter, I., Nowak, C., Nishimoto, T. et al. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398, 39–46 (1999). https://doi.org/10.1038/17969
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/17969
This article is cited by
-
Oncoprotein SET dynamically regulates cellular stress response through nucleocytoplasmic transport in breast cancer
Cell Biology and Toxicology (2023)
-
Nuclear Pore Dysfunction in Neurodegeneration
Neurotherapeutics (2022)
-
GEF-independent Ran activation shifts a fraction of the protein to the cytoplasm and promotes cell proliferation
Molecular Biomedicine (2020)
-
Functional analysis of an essential Ran-binding protein gene, CpRbp1, from the chestnut blight fungus Cryphonectria parasitica using heterokaryon rescue
Scientific Reports (2020)
-
The coming-of-age of nucleocytoplasmic transport in motor neuron disease and neurodegeneration
Cellular and Molecular Life Sciences (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.