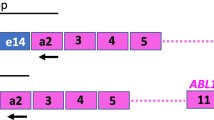
Abstract
We measured using a competitive quantitative polymerase chain reaction-capillary electrophoresis (PCR-CE)-based assay, the levels of bcr-abl transcripts in 44 patients with chronic myeloid leukemia (CML) after interferon-α (IFN-α) therapy, who achieved a major (10 patients, MCR group) or complete (34 patients, CCR group) cytogenetic response. All 34 CCR patients had molecular evidence of residual disease detected in bone marrow samples at the time of best karyotypic response. The median number of bcr-abl transcripts of 34 evaluable patients in the CCR group at the time of complete cytogenetic remission was 4/μg RNA (range 3–4600), while the median number of bcr-abl transcripts of 10 patients in the MCR group at the time of best cytogenetic response was 4490/μg RNA (range 600–23 900) (P = 0.000024). In nine CCR and five MCR patients we were able to quantify the amount of bcr-abl transcript both at diagnosis and after interferon therapy: no statistical difference (P = 0.18) was found between the two groups at diagnosis (median bcr-abl transcripts/μg RNA was 30 000 vs 39 650, respectively). During IFN-α therapy, the two groups were evaluable at the time of major karyotypic conversion: at this point, there was a statistical difference of expression of bcr-abl transcript between the CCR group (17 patients) (median 2700; range 76–40 000) and the MCR group (10 patients) (median 4490; range 600–23 900), respectively (P = 0.046). No differences of bcr-abl amount of transcript were found in patients with CCR obtained either by IFN-α therapy alone (20 patients) vs IFN-α plus ABMT (13 patients) (P = 0.47). We firstly demonstrated that although the CCR and MCR groups were clinically, cytogenetically and molecularly indistinguishable at diagnosis, the two groups could be recognized successfully during interferon therapy based on the level of bcr-abl transcript. Bone Marrow Transplantation (2000) 25, 729–736.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Rowley JD . A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining Nature 1973 24: 290–293
Nowell PC, Hungerford DA . A minute chromosome in human chronic granulocytic leukemia Science 1960 132: 1497–1499
Grosveld G, Verwoerd T, van Agthoven T et al. The chronic myelocytic cell line K562 contains a breakpoint in bcr and produces a chimeric bcr/c-abl transcript Mol Cell Biol 1986 6: 607–616
Shtivelman E, Gale RP, Dreazen O et al. Bcr-abl RNA in patients with chronic myelogenous leukemia Blood 1987 69: 971–973
Cross NCP, Lin F, Chase A et al. Competitive polymerase chain reaction to estimate the number of bcr-abl transcripts in chronic myeloid leukemia patients after bone marrow transplantation Blood 1993 82: 1929–1936
Chissoe SL, Bodenteich A, Wang YF et al. Sequence and analysis of the human ABL gene, the BCR gene, and regions involved in the Philadelphia chromosomal translocation Genomics 1995 27: 67–82
Lee MS, Kantarjian H, Talpaz M et al. Detection of minimal residual disease by polymerase chain reaction in Philadelphia chromosome-positive chronic myelogenous leukemia following interferon therapy Blood 1992 79: 1920–1923
Martinelli G, Testoni N, Montefusco V et al. Detection of bcr-abl transcript in chronic myelogenous leukemia patients by reverse-transcription-polymerase chain reaction and capillary electrophoresis Haematologica 1998 83: 593–601
Testoni N, Martinelli G, Farabegoli P et al. A new method of ‘in cell RT-PCR’ for the detection of bcr-abl transcript in chronic myeloid leukemia patients Blood 1996 87: 3822–3827
Lin F, Chase A, Bungey J et al. Correlation between the proportion of Philadelphia chromosome positive metaphases and levels of bcr-abl mRNA in chronic myeloid leukaemia Genes Chromos Cancer 1995 13: 110–114
Van Rhee F, Marks DI, Lin F et al. Quantification of residual disease in Philadelphia positive acute lymphoblastic leukemia: comparison of blood and bone marrow Leukemia 1995 9: 329–335
Hochhaus A, Lin F, Reiter A et al. Variable numbers of BCR-ABL transcripts persist in CML patients who achieve complete cytogenetic remission with interferon-α Br J Haematol 1995 91: 126–131
Malinge MC, Mahon FX, Delfau MH et al. Quantitative determination of the hybrid bcr-abl RNA in patients with chronic myelogenous leukemia under interferon therapy Br J Haematol 1992 82: 701–707
Miggiano MC, Gherlinzoni F, Rosti G et al. Autologous bone marrow transplantation in late first complete remission improves outcome in acute myelogenous leukemia Leukemia 1996 10: 402–409
The Italian Cooperative Study Group on Chronic Myeloid Leukemia . Interferon alfa-2a as compared with conventional chemotherapy for the treatment of chronic myeloid leukemia New Engl J Med 1994 330: 820–825
Mahon FX, Daheron L, Malinge MC et al. Polymerase chain reaction detection of residual disease in chronic myeloid leukemia patients in complete cytogenetic remission under interferon with or without chemotherapy Leukemia 1992 6: 1232–1234
Martiat P, Maisin D, Philippe M et al. Detection of residual BCR/ABL transcripts in chronic myeloid leukaemia patients in complete remission using the polymerase chain reaction and nested primers Br J Haematol 1990 75: 355–358
Opalka B, Kloke O, Bartram CR et al. Elimination by interferon-alpha of malignant clone in chronic myeloid leukaemia Lancet 1989 i: 1334 (letter)
Opalka B, Wandl UB, Becher R et al. Minimal residual disease in patients with chronic myelogenous leukemia undergoing long-term treatment with recombinant interferon α-2b alone or in combination with interferon γ Blood 1991 78: 2188–2193
Ozer H, George SL, Schiffer CA et al. Prolonged subcutaneous administration of recombinant α2b interferon in patients with previously untreated Philadelphia chromosome-positive chronic-phase chronic myelogenous leukemia: effect on remission duration and survival: Cancer and Leukemia Group B study 8583 Blood 1993 82: 2975–2984
Pardini S, Addis M, Dore F et al. Interferon-α2a therapy in CML: disappearance of BCR/ABL transcript in a case of long-lasting continuous cytogenetic conversion Haematologica 1994 79: 540–541 (review)
Talpaz M, Kantarjian H, Kurzrock R et al. Interferon alpha produces sustained cytogenetic responses in chronic myelogenous leukemia Ann Intern Med 1991 114: 532–538
Liberati AM, Donti E, Rosso C et al. Repeated PCR in CML during IFN-α therapy Eur J Haematol 1994 52: 152–155
Faderl S, Talpaz M, Kantarjian HM, Estrov Z . Should polymerase chain reaction analysis to detect minimal residual disease in patients with chronic myelogenous leukemia be used in clinical decision making? Blood 1999 93: 2755–2759
Goldman JM, Kaeda JS, Cross NCP et al. Clinical decision making in chronic myeloid leukemia based on polymerase chain reaction analysis of minimal residual disease Blood 1999 94: 1484–1486 (letter)
Pache G, Schmidt M, Vraetz T et al. An internally standardized competitive PCR Elisa for the precise quantification of clinical ex vivo gene transfer into hematopoietic stem cells Blood 1998 12: (Suppl.1) 150a (Abstr.605)
Wittor H, Emig M, Betzl G et al. Detection and quantification of bcr-abl transcripts using a novel one-step real-time RT-PCR approach with specific fluorescent hybridization probes Blood 1998 12: (Suppl.1) 73a (Abstr. 298)
Kafert S, Ganser A, Eder M . Differential quantification of alternatively spliced mRNAs using isoform specific taqman-probes and real-time RT-PCR Blood 1998 12: (Suppl.1) PAG (Abstr.3730)
Ladetto M, Donovan JW, Schlossman R et al. Limitations of quantitative detection of immunoglobulin heavy chain (IgH) gene rearrangements in multiple myeloma (MM) patients using real-time PCR Blood 1998 12: (Suppl.1) 258a (Abstr. 1056)
Seeger K, Kreuzer KA, Schmidt CA et al. Novel 5′ exonuclease-based real-time PCR assay for the detection of TEL-AML1 fusion in childhood acute lymphoblastic leukemia (ALL) Blood 1998 12: (Suppl. 1) 392a (Abstr.1622)
Bories D, Dumont V, Belhadj K et al. Real-time quantitative RT-PCR monitoring of chronic myelogenous leukemia Blood 1998 12: (Suppl.1) 73a (Abstr.296)
Shah MH, Yu H, Stover EH et al. Simultaneous quantification of cytokine gene expression in multiple organs during endotoxic shock Blood 1998 12: (Suppl.1) 545a (Abstr.2238)
Mensink E, Van de Locht A, Schattenberg E et al. Quantitation of minimal residual disease in Philadelphia chromosome positive chronic myeloid leukaemia patients using real-time quantitative RT-PCR Br J Haematol 1998 102: 768–774
Acknowledgements
The authors are grateful to Mr Robin MT Cooke for editorial assistance. This work was supported by Italian Association of Cancer Research (AIRC), by Italian CNR and by MURST 40% target projects and by ‘30 Ore per la Vita’ AIL grants.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Martinelli, G., Testoni, N., Amabile, M. et al. Quantification of BCR-ABL transcripts in CML patients in cytogenetic remission after interferon-α-based therapy. Bone Marrow Transplant 25, 729–736 (2000). https://doi.org/10.1038/sj.bmt.1702207
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bmt.1702207