Abstract
Despite widespread use of antidepressants, the factors underlying the behavioral response to antidepressants are unknown. It has been shown that antidepressant treatment promotes the proliferation and survival of neurons in the adult hippocampus via enhanced serotonergic signaling, but it is unclear whether hippocampal neurogenesis is responsible for the behavioral response to antidepressants. Furthermore, a large subpopulation of patients fails to respond to antidepressant treatment due to presumed underlying genetic factors. In the present study, we have used the phenotypic and genotypic variability of inbred mouse strains to show that there is a genetic component to both the behavioral and neuronal effects of chronic fluoxetine treatment, and that this antidepressant induces an increase in hippocampal cell proliferation only in the strains that also show a positive behavioral response to treatment. Furthermore, the behavioral and neuronal responses are associated with an upregulation of genes known to promote neuronal proliferation and survival. These results suggest that inherent genetic predisposition to increased serotonin-induced neurogenesis may be a determinant of antidepressant efficacy.
Similar content being viewed by others
INTRODUCTION
Major depressive disorder (MDD) affects up to 5% of the general population, with a further 15% suffering less severe forms of the disorder (Kessler et al, 2003). While 80% of patients will experience partial relief of depressive symptoms, complete remission occurs in only 50–60% of all cases despite treatment with multiple antidepressants (Nestler et al, 2002; Rush et al, 2006). Even in patients who respond to drug therapy, chronic treatment—on the order of weeks or months—is required before antidepressants provide clinical benefit. The result is that many MDD patients suffer for months or years before finding effective treatment. It is likely that individual genetic differences regulate the response to antidepressants, but few polymorphisms have been identified (Binder and Holsboer, 2006; McMahon et al, 2006).
One barrier to developing more effective treatments for depression is that the mechanism by which antidepressants act is poorly understood: although antidepressants produce elevation of synaptic serotonin levels within minutes of administration (Ross and Renyi, 1975; Bel and Artigas, 1992; Malagie et al, 1995), measurable clinical response may not occur for weeks (Katz et al, 2004). It has, therefore, been hypothesized that increased monoaminergic transmission is a precursor to molecular and neural changes that underlie the remission of depression. These changes are likely to include chronic alterations in gene expression and neuronal plasticity that change the activity of the neural circuitry that regulates mood (Nestler et al, 2002). One of the most intriguing hypotheses associates antidepressant treatment with elevated neuronal proliferation and survival rates (Duman et al, 1997; Altar, 1999; Duman et al, 2001).
Neurogenesis is primarily defined as the production of new nervous tissue, and is characterized by labeling cells undergoing S phase with the nucleotide precursors bromodeoxyuridine (BrdU) or [3H]thymidine (Duman et al, 2001). However, the definition of neurogenesis may be generalized to encompass subsequent related processes, such as neuronal differentiation, survival, and incorporation into functional networks. In the adult mammalian brain, neurogenesis is largely restricted to the olfactory bulbs and the subgranular zone (SGZ) of the hippocampus, a region lined with serotonergic fibers (Kaplan and Hinds, 1977). Approximately 250 000 new neurons are born monthly in the SGZ of the adult rat, 50–70% of which differentiate into neurons that migrate into the granule cell layer (Hastings and Gould, 1999; Cameron and McKay, 2001). Antidepressants have been shown to increase both proliferation and survival of hippocampal neurons under basal conditions: in the hippocampus of unstressed mice, chronic antidepressant treatment results in a 60% increase in BrdU labeling (Santarelli et al, 2003). Antidepressants can also ameliorate the negative impact of stressors or glucocorticoids on neuronal proliferation: in rats, administration of fluoxetine for 1 week following an acute stressor prevents the stress-induced reduction in neuronal proliferation (Malberg and Duman, 2003), and in tree shrews, antidepressant administration prevents the effects of chronic stress on neuronal atrophy (Czeh et al, 2001). The time course of antidepressant-induced changes in neurogenesis closely mimics the time required for mood-elevating effects in humans: 1–5 days of selective serotonin reuptake inhibitor (SSRI) treatment has no effect on BrdU labeling, 7–14 days of treatment increases the rate of neuronal proliferation, but a minimum of 3–4 weeks of treatment is required to produce an increase in survival (Malberg et al, 2000; De Foubert et al, 2004). It has recently been shown that antidepressant-induced neurogenesis may be necessary for the behavioral response to antidepressants: mice that received chronic fluoxetine treatment in combination with irradiation of the hippocampus to prevent neurogenesis failed to show a response to treatment in two separate behavioral tasks (Santarelli et al, 2003). To date, no human studies have conclusively associated a change in neurogenesis with depression or the response to antidepressants. However, a number of imaging studies have shown that depressed patients have reduced hippocampal volume compared to normal subjects (Sheline et al, 1996; Videbech and Avnkilde, 2004), while depressed patients treated successfully with antidepressants have a normal hippocampal volume (Sheline et al, 2003).
The effect of antidepressants on neurogenesis is primarily mediated by serotonergic regulation of intracellular signaling mechanisms that upregulate transcription and growth factors involved in neuronal proliferation (Duman et al, 2001). In general, conditions that are associated with reduced serotonergic transmission, such as aging, malnutrition, or chronically elevated corticosterone reduce SGZ proliferation, while stimuli that increase neurogenesis, such as electroconvulsive therapy and adrenalectomy also increase serotonin levels (Gould, 1999). Under basal conditions, serotonin appears to play a vital role in neurogenesis: injection of either a serotonin neuron-specific neurotoxin or an inhibitor of serotonin synthesis into the raphe nucleus results in a drastic decrease in BrdU labeling (Brezun and Daszuta, 1999; Banasr et al, 2004). Serotonergic signaling activates cAMP/cAMP response element-binding protein (CREB), which upregulates expression of Bdnf, a potent inducer of neurogenesis (Nibuya et al, 1996). These results suggest that the ultimate effect of the antidepressant-induced increase in serotonin is to increase neurotrophins, which may be the primary mediators of the mood-elevating effects of antidepressants.
In the present study, we exploited the genotypic and phenotypic differences among inbred strains of mice to gather evidence supporting the association between the stimulation of hippocampal cell proliferation, as measured by BrdU labeling, and antidepressant efficacy, which was measured using the tail suspension test (TST). We also compared transcriptome-wide hippocampal gene expression differences between control mice and mice treated chronically with antidepressants to identify the genes regulated by antidepressant treatment.
MATERIALS AND METHODS
Behavioral Testing
Male 129S1/SvImJ, A/J, DBA/2J, and SWR/J mice, age 3–5 weeks, were obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained on a 12 : 12 light/dark cycle with lights on at 0700 h. Following at least 1 week of acclimation, mice were provided with either untreated or fluoxetine-treated drinking water available ad libitum. Fluoxetine-HCl was obtained from Spectrum Chemicals (Gardena, CA). Differences in daily water consumption were previously determined for each strain by measuring water intake for 3 weeks from at least 12 mice per strain (129S1/SvImJ and A/J mice=5.2 ml (±0.36 and 0.22 ml, respectively) per day, SWR/J=5.9 ml (±0.29 ml) per day, and DBA/2J=6.4 ml (±0.58 ml) per day). We also determined the average weight at 10 weeks of age (129S1/SvImJ=23.9±2.8 g, A/J=24.8±2.3 g, DBA/2J=25.5±1.4 g, SWR/J=20.8±1.3 g), and used this information to dilute fluoxetine in the water to provide a daily dosage of 0, 5, 10, 14, or 18 mg/kg/mouse, depending on the experimental context. To determine whether the addition of fluoxetine to the water-affected drinking behavior, and, therefore, fluoxetine intake, water intake for fluoxetine-treated mice was monitored. No difference in water intake between the control and drug-treated mice was observed.
Mice were 8–12 weeks of age at the time of behavioral testing, which was performed during the light phase of the light/dark cycle between 1300 and 1600 h. Behavioral despair was measured using a mouse tail suspension apparatus from Med Associates (Georgia, VT). Scotch tape was used to suspend mice by the tail inside the cubicle, and activity, as measured by load, was recorded for 7 min in 60-s blocks, with threshold=3, gain=8, and resolution=200 ms. As all strains were uniformly active for the first minute, only minutes 2–7 were used for determining the percent of time spent immobile, which was calculated by dividing the number of seconds of immobility from blocks 2 to 7 by 360. All results are expressed in terms of the percent of time spent immobile. Approximately 20% of DBA/2J mice climbed their tails during behavioral testing; these mice were excluded from data analysis. All animal testing was conducted under TSRI-Scripps Florida IACUC-approved protocols.
Dose–Response
To determine the most efficacious dose of fluoxetine, DBA/2J mice were administered fluoxetine-treated drinking water at a daily dose of 0, 5, 10, 14, or 18 mg/kg/mouse for 1, 6, or 21 days before behavioral testing. A total of 24 mice per dose were used for the 6- and 21-day time points, and 12 mice per dose were used for the 1-day time point. Fluoxetine treatment was timed so that all mice were tested over a 3-day period, with all doses and treatment durations represented equally on each day of testing. Immediately following the TST, mice were euthanized by rapid decapitation and trunk blood was collected for serum fluoxetine and norfluoxetine measurements. Blood was allowed to clot on ice, centrifuged, and serum was retrieved and stored at −20°C until analysis by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS). Hippocampal tissue was also collected from twelve 21-day, 18 mg/kg and twelve 21-day, 0 mg/kg mice for microarray analysis. Immediately following decapitation, brains were placed in a corornal brain block (Stoelting Co., Wood Dale, IL), and a 2 mm section extending from approximately Bregma −1.7 to Bregma −3.7 was removed. The hippocampus was dissected away from the cortex and midbrain, placed in 500 μl TRIzol (Invitrogen, Carlsbad, CA), and stored at −80°C until RNA extraction.
Cell Proliferation
Ten to twelve 129S1/SvImJ, A/J, DBA/2J, and SWR/J mice were administered fluoxetine-treated water at a dose of 0 or 18 mg/kg/mouse for 21 days. On the 20th day of treatment, mice received a single intraperitoneal injection of 200 mg/kg BrdU (Sigma-Aldrich, St Louis, MO). Twenty-four hours after the BrdU injection, behavior was assessed in the TST. Immediately following the TST, mice were anesthetized with ketamine/xylazine, approximately 100 μl of blood was collected from the submandibular vein, and mice were transcardially perfused with 1% paraformaldehyde for 1 min followed by 4% paraformaldehyde for 7 min. Brains were removed, postfixed overnight in 4% paraformaldehyde at 4°C, cryoprotected in 30% sucrose/phosphate-buffered saline (PBS, pH 7.2) for 24–48 h, and stored at −80°C until sectioning.
To determine the fate of BrdU-labeled cells, 10 DBA/2J mice were treated with fluoxetine as described above. On the 21st day of fluoxetine treatment, the mice received two injections of BrdU (200 mg/kg i.p.) over the course of 6 h. Seventy-two hours after the second BrdU injection, the mice were euthanized by perfusion.
A freezing microtome was used to take 40 μm sections through the hippocampus from Bregma −0.94 to Bregma −4.04 mm. The sections were subjected to epitope retrieval (50% formamide/2 × SSC at 65°C for 2 h), washed in 2 × SSC at 37°C, and treated with 2N HCl at 37°C for 30 min, followed by a 10-min wash in 0.1 M borate buffer at room temperature. Sections were pretreated with 5% normal rabbit serum, 3% bovine serum albumin (BSA), and 0.1% Triton for 1 h. Sections from mice receiving a single BrdU injection were then incubated in rat monoclonal anti-BrdU (1 : 200, Oxford Biotechnology, Oxfordshire, UK) while mice receiving the dual BrdU injections were incubated in anti-BrdU and biotinylated mouse monoclonal anti-NeuN (Chemicon, Temencula, CA), 3% normal rabbit serum, and 0.1% Tween-20 overnight at 4°C. BrdU staining was visualized using rabbit anti-rat fluorescein isothiocyanate (FITC) (1 : 200, Vector Labs, Burlingame, CA) and NeuN staining was visualized using avidin Texas Red (1 : 400, Vector Labs). For the mice euthanized 24 h after BrdU injection, the number of BrdU-positive cells in the dentate gyrus (DG) was determined for each mouse by counting BrdU-positive cells along the SGZ from every sixth brain section and dividing the total number of cells counted by the number of sections counted. For the mice euthanized 72 h after BrdU treatment, all BrdU cells were counted and examined for colocalization with NeuN fluorescence. Cell counting was performed on an Olympus BX61 by an investigator blind to experimental conditions.
Determination of Serum Levels of Fluoxetine and Norfluoxetine
Serum levels of fluoxetine and norfluoxetine were quantitated by LC-MS/MS using a standard curve between 20 and 10 000 ng/ml. A total of 10 μl serum was treated with 150 μl acetonitrile containing 5 μg/ml of each internal standard, fluoxetine-D6 and norfluoxetine-D6, and samples were filtered through a 0.45 μm filter plate. Fifty microliters of the filtrate was diluted in 0.1% formic acid and analyzed on a Sciex 4000 LC-MS/MS (Sciex Inc., Concord, ON) equipped with a Waters YMC Cyano HPLC column (2.0 × 23 mm) (Milford, MA) using a mobile phase of water/acetonitrile/formic acid (75 : 25 : 0.1). The peak areas of the m/z 310 → 44 fluoxetine and m/z 296 → 134 norfluoxetine ions were measured against the peak areas of the m/z 316 → 44 fluoxetine-D6 and m/z 302 → 140 fluoxetine-D6 ions of the internal standards. Quantitation was performed using separate weighted linear least-squares regression analyses generated from fortified plasma calibration samples.
Microarray
Individual bilateral hippocampi from DBA/2J mice receiving 0 or 18 mg/kg/day of fluoxetine for 21 days were homogenized in 500 μl TRIzol using a QIAgen TissueLyser (15 min at 30 Hz, QIAgen, Valencia, CA). RNA was extracted by phenol–chloroform phase separation and further processed using the RNAeasy miniprep kit (QIAgen). For each treatment group of 12 animals, separate RNA pools were made using hippocampal RNA from six mice per pool. These biological replicates were processed separately through the entirety of the microarray procedure. One microgram of total RNA from each pool was used as a template to synthesize complementary DNA (cDNA) and biotinylated cRNA (Enzo kit, Affymetrix, Santa Clara, CA) using standard Affymetrix protocols. cRNA was hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 arrays. Data were analyzed using ArrayAssist software (Stratagene, La Jolla, CA) and normalized with the GCRMA algorithm. A probeset had to obtain a minimal intensity value of 100 to be considered as detecting expression above background. Therefore, any probeset that failed to meet this cutoff was removed from the analysis. Ratios of intensity differences were generated and genes showing at least a twofold difference between experimental conditions were identified for further analysis. Data from the microarrays can be retrieved from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession #GSE6476.
Real-Time PCR
Male A/J and DBA/2J mice were provided with untreated (six mice per strain) or fluoxetine-treated (six mice per strain, 18 mg/kg/day) drinking water for 3 weeks. At the end of the treatment period, bilateral hippocampi were removed and RNA was extracted as described above. RNA samples were pooled, reverse-transcribed using the Superscript III kit (Invitrogen), and used for real-time PCR gene expression analysis of 15 genes identified as significantly upregulated in fluoxetine-treated DBA/2J hippocampus by microarray analysis. Real-time PCR (reverse transcription (RT)-PCR) was performed on 100 ng of cDNA using SYBR Green iMix (Biorad, Hercules, CA) and primers designed over exon–intron boundaries (IDT, Coralville, IA). The reactions were assayed on an ABI 7900 HT Real-Time PCR system (Applied Biosystems, Foster City, CA). Samples were run in triplicate with a Gapdh control (QIAgen) on each plate and the results were analyzed with the comparative CT method using SDS software v2.2.2 (Applied Biosystems).
Statistical Analysis
Tail suspension and serum fluoxetine data from the dose–response study were analyzed by two-way analysis of variance (ANOVA) (dose, duration) and Student's t-test (post hoc analysis) using the JMP statistical software package (SAS Institute, Cary, NC). The effect of strain on TST immobility and BrdU labeling in control mice was analyzed by one-way ANOVA in JMP, and the within-strain effect of fluoxetine on tail suspension behavior and hippocampal cell proliferation was analyzed by t-test (two-tailed unequal variance) in Excel (Microsoft, Redmond, WA). The effect of fluoxetine on hippocampal gene expression was analyzed by calculating fold change in Excel, and significantly enriched Gene ontology (GO) categories were identified using L2L software (Newman and Weiner, 2005).
RESULTS
Dose Response
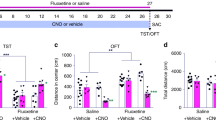
To determine the most appropriate dose of fluoxetine, a commonly used SSRI, for chronic oral treatment, we performed a fluoxetine dose–response test using DBA/2J mice, a strain that is behaviorally responsive to fluoxetine (Lucki et al, 2001). Mice were treated for 1 (acute), 6 (subchronic), or 21 days (chronic) with 0, 5, 10, 14, or 18 mg/kg fluoxetine provided ad libitum in the drinking water. At the end of the treatment period, the effect of fluoxetine dose and duration were evaluated in the TST. This test measures the duration that a rodent will persist in escape-oriented behavior when faced with an inescapable stress. The TST has been shown to predict the efficacy of known antidepressants, and is commonly employed in detecting new antidepressants that have a novel mechanism of action (Steru et al, 1985; Chaki et al, 2004). In the majority of cases, antidepressants increase the duration an animal will struggle and reduce the time spent immobile when suspended in the TST, a state best described as behavioral despair (Cryan et al, 2005).
There were significant main effects of both drug dose and treatment duration (both p<0.001), and a significant dose × duration interaction (p=0.001, Figure 1). Post hoc analysis showed that after 1 day of treatment, none of the doses had any significant effect on the percent of time mice spent immobile in the TST. After 6 days of treatment, both the 14 and the 18 mg/kg doses produced a significant reduction in immobility: mice in these groups averaged 5% immobility, compared to 14% immobility in the control group. After 21 days of treatment, the 18 mg/kg dose reduced immobility to the same statistical extent as the 6-day, 14- and 18 mg/kg treatments. While the 14 mg/kg dose also significantly reduced immobility, its effect was significantly different (p<0.05) from the 21-day, 18 mg/kg and 6-day, 14- and 18 mg/kg doses.
The lack of a behavioral effect of 1 day of oral fluoxetine treatment in the present study is in contrast to the majority of studies involving antidepressant treatment, most likely because investigators have traditionally measured behavior 30 min after an i.p. injection of antidepressants (Porsolt et al, 1977; Lucki et al, 2001; Cryan et al, 2005). However, these results are in agreement with data from other labs that have delivered fluoxetine orally. Dulawa et al (2004) found that 4-day fluoxetine treatment had no effect on either the open field test, a measure of general activity and anxiety-like behavior, or the forced swim test (FST), another measure of behavioral despair. Similarly, Caldarone et al (2003) found that 21 days of treatment with the tricyclic antidepressant amitriptylene reduced FST immobility to a greater extent than 4 days of treatment. Since chronic antidepressant treatment is required for clinical efficacy in humans, these results suggest that, in mice, oral dosing of antidepressants more closely resembles human treatment paradigms than the acute injection model. Furthermore, oral dosing avoids the inevitable handling stress and pain of daily injections that would likely impact both the behavioral and neurological phenotypes.
Serum was collected from 12 mice per dose/duration group to measure serum fluoxetine levels. There were significant dose, duration, and dose × duration effects (all p<0.001, Table 1). Post hoc analysis showed that 1 day of treatment resulted in low, but dose-dependent, serum fluoxetine levels; by 6 days of treatment, average serum fluoxetine levels were near those observed after 21 days of treatment. Because there was large within-group variability in serum fluoxetine levels, we used Pearson's r-test to determine whether there was an association between individual serum fluoxetine and TST performance. No association was found (r=0.18). This is consistent with human studies, which have found no significant correlation between serum fluoxetine or norfluoxetine levels and antidepressant efficacy (Beasley et al, 1990; Koran et al, 1996; Amsterdam et al, 1997).
Strain-Dependent Effects of Chronic Fluoxetine Treatment on Behavior
To determine whether there was a genetic component to baseline TST immobility and the behavioral response to chronic antidepressant treatment, mice from four inbred strains—129S1/SvImJ, A/J, DBA/2J, and SWR/J—were treated chronically with 0 or 18 mg/kg fluoxetine and then tested in the TST (Figure 2a). In the control mice, we observed strain differences (p<0.01) in the percent of time spent immobile: DBA/2J mice were immobile for approximately 17% of the test (n=4), SWR/J for 21% (n=6), 129S1 for 30% (n=12), and A/J for 43% (n=5). Analysis of the within-strain effect of fluoxetine on TST immobility showed that, in DBA/2J (n=5) and 129S1/SvImJ mice (n=12), chronic fluoxetine significantly reduced the TST immobility (p⩽0.01). In A/J mice (n=5), chronic fluoxetine had no effect on immobility, while in SWR/J mice (n=6), fluoxetine treatment significantly increased the immobility (p=0.01).
Association between Antidepressant Efficacy and Hippocampal Cell Proliferation
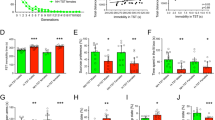
Control and fluoxetine-treated mice were euthanized 24 h after a single BrdU injection. The rate of cell proliferation in control and fluoxetine-treated mice was determined by counting BrdU-positive cells throughout the DG (Kempermann et al, 1997). Labeled cells were primarily restricted to the SGZ, although labeled cells were occasionally observed in the hilar region. Consistent with the estimated adult neuronal cell cycle period of 24–25 h (Cameron and McKay, 2001), labeled cells were often observed in groups of 2 or 4, indicating that the cells labeled at the time of the BrdU injection had had an opportunity to divide once since the initial labeling period. BrdU-positive cells were not observed in either the cortex or the midbrain.
There was a significant effect of strain (p<0.001) on basal cell proliferation as measured by the average number of BrdU-labeled cells per section: 129S1 mice had the fewest labeled cells, DBA/2J and SWR/J mice exhibited intermediate cell proliferation, and A/J mice exhibited the highest rate of cell proliferation (Figure 2b). The difference between DBA/2J and A/J is similar to that observed in previous studies, which have shown that A/J mice exhibit a higher rate of hippocampal cell proliferation than DBA/2J mice (Kempermann et al, 2006). In two strains, DBA/2J and 129S1, chronic fluoxetine treatment increased the number of BrdU-positive cells (p=0.016 and 0.02, respectively). Significantly, both of these strains also exhibited a positive behavioral response to chronic fluoxetine treatment. In contrast, fluoxetine treatment had no effect on the number of labeled cells in the A/J and SWR/J strains, both of which did not show a decrease in TST immobility following fluoxetine treatment (Figure 3).
BrdU labeling (arrows) in the dentate gyrus of the hippocampus in control (a, b) and fluoxetine-treated (c, d) SWR/J and 129S1 mice. Chronic fluoxetine treatment significantly increased BrdU labeling in the hippocampus of 129S1 mice, but had no effect on BrdU labeling in SWR/J mice. Mol=molecular layer, gcl=granule cell layer, × 20.
To confirm that BrdU labeling is a proxy for neurogenesis, DBA/2J mice were treated with fluoxetine for 3 weeks and euthanized 72 h after BrdU treatment. This time period is associated with a high level of neuronal differentiation and represents the peak time for colocalization of the mature neuronal marker NeuN with BrdU-labeled cells (Kempermann et al, 2004; Steiner et al, 2004). A total of 2276 BrdU-positive neurons were counted from the DG of 10 mice; of these, 1374 (58%) were also positive for NeuN (Figure 4). These results are consistent with previous reports of the percent of BrdU-labeled cells that ultimately mature into neurons (Minturn et al, 1995; Cameron and McKay, 2001; Mignone et al, 2004).
Brains from bromodeoxyuridine (BrdU)-treated DBA/2J mice were stained for BrdU (green, a, d) and the mature neuronal marker NeuN (red, b, e). Examples of a BrdU-positive/NeuN-negative cell (c) and a BrdU-positive/NeuN-positive cell (f) are shown. All photomicrographs were taken using a 60 × objective.
Effect of Chronic Fluoxetine Treatment on Hippocampal Gene Expression
To produce a more complete picture of the molecular changes that occur in the hippocampus following chronic antidepressant treatment, we used a whole-genome microarray to evaluate gene expression in hippocampi from DBA/2J mice, the strain that showed the strongest antidepressant-induced change in both behavior and hippocampal cell proliferation. Two biological replicates per treatment group (0 or 18 mg/kg fluoxetine) were hybridized to Affymetrix Mouse Genome 430 2.0 microarrays. Following normalization with the GCRMA algorithm, the biological replicates were averaged and fold change was determined for each gene by dividing the intensity of expression in the fluoxetine group by the intensity of expression in the control group. Genes with a fold change >2.0 or <0.5 were considered significantly up- or downregulated. In total, 83 unique genes were upregulated and 17 genes were downregulated by chronic fluoxetine treatment (Table 2, Supplementary Table S1). Most highly upregulated were Preproenkephalin 1, a possible quantitative trait gene for bipolar disorder (Ogden et al, 2004), and Galanin, antagonists of which prevent the behavioral effect of fluoxetine in the TST and the FST (Holmes et al, 2005; Swanson et al, 2005).
To validate the microarray results as well as determine whether the transcriptional changes observed were specific to a fluoxetine-responsive strain, we quantitated the expression of a subset of genes identified by microarray in independent hippocampal tissue samples from DBA/2J and A/J mice, a nonresponder strain, by RT-PCR. Of the 15 genes analyzed by RT-PCR, all but one (Lgals1) were significantly upregulated in fluoxetine-treated DBA/2J mice compared to control DBA/2J mice. In contrast, none of the 15 genes were affected by fluoxetine treatment in A/J mice (Figure 5).
RT-PCR analysis of control and fluoxetine-treated DBA/2J hippocampus confirmed microarray results. Of the 14 genes shown to be upregulated in fluoxetine-treated DBA/2J hippocampus compared to control hippocampus, none were upregulated in fluoxetine-treated A/J hippocampus relative to control. *p⩽0.01.
DISCUSSION
Recent studies have shown that inbred mouse strains vary in the rate of adult neuronal proliferation and survival, and in the behavioral response to antidepressant treatment (Lucki et al, 2001; Kempermann et al, 2006). Given the strain-dependent variability in both antidepressant response and neurogenesis, we hypothesized that there might be a genetic component to the effect of fluoxetine on neuronal proliferation. Furthermore, a correlation between the strain-dependent effect of fluoxetine on neurogenesis and the strain-dependent behavioral efficacy of fluoxetine would provide strong evidence in support of the theory that antidepressant-induced neurogenesis plays a role in the behavioral response to antidepressants. We tested this hypothesis using four inbred strains of mice treated chronically with fluoxetine. Significantly, the two strains, 129S1/SvImJ and DBA/2J, that exhibited a positive behavioral response to fluoxetine treatment were the same strains that responded to fluoxetine treatment with an increase in hippocampal cell proliferation. Double-labeling studies showed that almost 60% of BrdU-labeled cells expressed the mature neuronal marker NeuN 3 days after labeling, indicating that the majority of the new cells were destined to become neurons. These results suggest that the strain-dependent effect of fluoxetine treatment on the rate of hippocampal cell proliferation is associated with the behavioral response to fluoxetine.
A number of researchers have proposed a causative link between neurogenesis and antidepressant efficacy, but only a limited number of studies have demonstrated an association between antidepressant-induced increase in neurogenesis and a behavioral effect. Most notably, Santarelli et al (2003) showed that inhibiting hippocampal proliferation by targeted X-ray irradiation of hippocampal precursor cells prevented the behavioral effects of fluoxetine on two behavioral measures of antidepressant efficacy. While control experiments suggested that irradiation did not disrupt hippocampal function beyond limiting proliferation, the possibility remains that unknown molecular factors important to antidepressant efficacy and/or hippocampal function were affected by the procedure. X-ray irradiation of the hippocampus has been shown to cause cognitive deficits in mice (Rola et al, 2004), and may increase apoptosis, produce changes in genes associated with DNA damage and stress response, alter the morphology or functionality of mature neurons, and alter blood flow to irradiated regions (Gobbel et al, 1998; Shirai et al, 2006). Therefore, there remains a need for less invasive methods to test the link between neurogenesis and antidepressant efficacy. In the present study, a genetic model was utilized for the investigation of the role of increased hippocampal cell proliferation in antidepressant efficacy. The availability of a large number of inbred strains will allow for the replication of our finding on a more comprehensive scale.
The inbred strain model may also be expanded upon to dissect the effect of antidepressants on additional aspects of neurogenesis (Kempermann et al, 2006). Although the present study addressed only neuronal proliferation, there is evidence that antidepressant treatment has an effect on other phases of neurogenesis as well. Imipramine and fluoxetine have been shown to increase both neuronal proliferation and survival in the adult hippocampus via activation of trkB/BDNF signaling: mice deficient in BDNF do not show enhanced neuronal survival after antidepressant treatment (Sairanen et al, 2005). Fluoxetine also promotes plasticity in the hippocampus by increasing pyramidal neuron dendritic spine density (Hajszan et al, 2005), which may provide a general enhancement of the synaptic input that new neurons require for survival and network integration (Kee et al, 2007; Toni et al, 2007).
In the present study, A/J mice exhibited higher baseline new neuron formation than DBA/2J mice, a finding in agreement with previous reports (Kempermann and Gage, 2002; Kempermann et al, 2006). Although it is possible that A/J mice failed to show an increase in hippocampal cell proliferation in response to fluoxetine because of a ceiling effect (eg baseline proliferation is already at maximum), this is an unlikely interpretation: C57BL/6J (B6) mice have been shown to have a higher rate of hippocampal cell proliferation than A/J mice, but, unlike A/J, B6 mice are behaviorally responsive to the antidepressant effects of fluoxetine treatment (Kempermann et al, 1997, 2006; Bai et al, 2001; Hayes and Nowakowski, 2002). It is important to note, however, that our data suggest that the rate of baseline hippocampal cell proliferation is not correlated with baseline TST immobility: rather, the correlation between cell proliferation and TST activity is specific to the effect of fluoxetine.
With regard to behavioral performance, we observed a statistically significant strain effect on immobility in the TST, with DBA and SWR mice representing low baseline immobility and A/J and 129S1 mice representing high baseline immobility. However, there was no apparent association between baseline immobility and the behavioral response to antidepressants, as one strain from each group (low vs high immobility) exhibited a behavioral response to fluoxetine. Despite concerns that the effect of environment may outweigh the genetic effects in the measurement and interpretation of behavioral phenotypes like those presented here (Crabbe et al, 1999; Wahlsten et al, 2003), our results regarding the effect of strain and fluoxetine treatment on TST performance are consistent with those from other groups. Both Crowley (Crowley et al, 2005) and Trullas (Trullas et al, 1989) ranked A/J mice as high immobility compared to DBA/2J mice, as did Liu (Liu and Gershenfeld, 2003), although the latter group observed only minimal differences between strains. Similarly, both Crowley and Liu observed a significant effect of antidepressant (imipramine and citalopram, respectively) treatment on DBA/2J, but not A/J or SWR/J, as measured by TST performance (Liu and Gershenfeld, 2001). The observed increase in SWR/J immobility following fluoxetine treatment in the present study is surprising but not completely unprecedented: Liu and Gershenfeld (2003) and Cryan et al (2005) found that AKR mice exhibit a similar increase in immobility following imipramine treatment. In all cases, the raw values for the various phenotypes differ among labs, a phenomenon most likely due to a combination of factors, including the method of measurement (manual vs automated), handling, housing, and the type and duration of drug treatment (Crabbe et al, 1999). However, the preservation of the relative order of the inbred strains for these quantitative values indicates an overriding genetic control that can reliably be used to further dissect this biology.
The variation in the manner in which different mouse strains respond to fluoxetine resembles the variation in human responses to antidepressant treatment: complete remission of depression occurs in only 50–60% of patients, and the clinical efficacy of any given antidepressant varies greatly among individuals (Rush et al, 2006). Polymorphisms in several genes involved in monoaminergic signaling have been linked to antidepressant efficacy, but it is likely that that many other genes underlie individual differences in antidepressant response (Serretti et al, 2005; Binder and Holsboer, 2006). Inbred mouse strain genomics provides an alternate means to identify these genes. A form of haplotype association mapping (Pletcher et al, 2004) has recently been used to identify genes responsible for variation in complex biologies such as drug metabolism (Guo et al, 2006), tumor susceptibility (Liu et al, 2006), and liver fibrosis (Hillebrandt et al, 2005). The strain-dependent differences in baseline TST immobility and the behavioral response to antidepressants suggest that, provided a larger strain set, these traits will also be amenable to dissection by haplotype mapping. By examining genetic and genomic differences between responder and nonresponder strains, we may be able to identify new antidepressant targets.
As an initial step in identifying genes involved in the response to chronic fluoxetine treatment, we analyzed expression of ∼39 000 transcripts in hippocampi from control and fluoxetine-treated DBA/2J mice. We identified 83 unique genes upregulated and 17 genes downregulated in the brains of fluoxetine-treated mice. A number of these genes, including Bdnf, Npy, Egr3, and Cox2, have also been shown to be induced by electroconvulsive seizures, another type of antidepressant treatment (Newton et al, 2003). It is likely that the upregulated genes are associated specifically with the behavioral and/or neuronal response to treatment, as none of 15 genes measured were affected by fluoxetine treatment in A/J mice, a strain that does not show a behavioral or neuronal response to fluoxetine. Therefore, these genes represent potentially novel therapeutic targets for antidepressant treatments.
Biological relevance of the microarray data was assessed using L2L software, which takes into account the total number of probes targeting individual GO categories and compares the number of matches expected by chance for each GO category to the actual number of matches (Table 3) (Newman and Weiner, 2005). Not surprisingly, a significant majority of upregulated genes were associated with neurogenesis and/or synaptogenesis: these include factors known to promote the proliferation of neuronal progenitor cells (Npy, Edg3, Lgals1, Serpinf1/Pedf, Hspb1), growth factor-associated genes involved in survival and differentiation of adult neuronal cells (Bdnf, Inhba, Acvr1c, c-Ret), and downstream immediate early genes (IEGs) and transcription factors (c-fos, Sox11, Egr3, S100a6). Among the downregulated genes were Calb1, a marker of mature granule neurons, Prdm5, which has growth-suppressive effects, and desmoplakin, which promotes cell adhesion and is, therefore, likely to reduce proliferation. Although published reports of global transcriptome and proteome analysis following chronic antidepressant treatment are limited, several other groups have also observed an upregulation of neurogenic factors following treatment with a wide variety of antidepressants including fluoxetine and venlafaxine (Khawaja et al, 2004) and amitriptyline, moclobemide, and clorgyline (Drigues et al, 2003).
The present microarray results support the hypothesis that BDNF and other growth factors play an important role in antidepressant activity. Bdnf, which is fivefold upregulated in the hippocampii of fluoxetine-treated mice compared to controls, is a downstream target of the CREB pathway, which is activated by 5HT1A signaling (Nibuya et al, 1996; Dowlatshahi et al, 1998; Santarelli et al, 2003). CREB has been shown to be responsible for upregulating Bdnf expression following chronic antidepressant treatment (Conti et al, 2002); BDNF signaling via the trkB receptor mediates the effect of chronic antidepressant treatment on neuronal survival in the hippocampus and on performance in a behavioral despair task (Saarelainen et al, 2003; Sairanen et al, 2005). When administered centrally, BDNF has been shown to have antidepressant-like effects on behavioral tasks (Hoshaw et al, 2005).
The upregulation of Bdnf, along with genes essential for FGF and GDNF signaling, by fluoxetine treatment suggests that the effect of antidepressants on neurogenesis is mediated by serotonergic regulation of intracellular signaling mechanisms that upregulate transcription and growth factors involved in neuronal proliferation (Gould, 1999). Growth factors act via MAPK signaling to induce the transcription of IEGs and transcription factors that promote the proliferation of neuronal progenitor cells and the survival and differentiation of newly born neurons. A recent study has identified a specific class of neuronal precursor cells, amplifying neural progenitors that appear to be sensitive to chronic fluoxetine treatment (Encinas et al, 2006); it is possible that the strain-dependent effect of fluoxetine on neurogenesis is due to genetic polymorphisms associated with this type of cell.
Our findings indicate that both the behavioral and neuronal responses to fluoxetine are under genetic control, and support the hypothesis that increased hippocampal cell proliferation plays a role in the behavioral response to fluoxetine. Future work will seek to expand the range of antidepressants used and explore the effects of these drugs on other aspects of neurogenesis, including neuronal survival and pruning, to better understand the genetic factors that contribute to individual differences in antidepressant response. This work also provides new mouse models for the further dissection of antidepressant efficacy and mechanism. Evaluating the effects of chronic antidepressant treatment in a larger number of inbred mouse strains may provide a way to identify novel genes that regulate antidepressant efficacy.
Accession codes
References
Altar CA (1999). Neurotrophins and depression. Trends Pharmacol Sci 20: 59–61.
Amsterdam JD, Fawcett J, Quitkin FM, Reimherr FW, Rosenbaum JF, Michelson D et al (1997). Fluoxetine and norfluoxetine plasma concentrations in major depression: a multicenter study. Am J Psychiatry 154: 963–969.
Bai F, Li X, Clay M, Lindstrom T, Skolnick P (2001). Intra- and interstrain differences in models of behavioral despair. Pharmacol Biochem Behav 70: 187–192.
Banasr M, Hery M, Printemps R, Daszuta A (2004). Serotonin-induced increases in adult cell proliferation and neurogenesis are mediated through different and common 5-HT receptor subtypes in the dentate gyrus and the subventricular zone. Neuropsychopharmacology 29: 450–460.
Beasley Jr CM, Bosomworth JC, Wernicke JF (1990). Fluoxetine: relationships among dose response adverse events and plasma concentrations in the treatment of depression. Psychopharmacol Bull 26: 18–24.
Bel N, Artigas F (1992). Fluvoxamine preferentially increases extracellular 5-hydroxytryptamine in the raphe nuclei: an in vivo microdialysis study. Eur J Pharmacol 229: 101–103.
Binder EB, Holsboer F (2006). Pharmacogenomics and antidepressant drugs. Ann Med 38: 82–94.
Brezun JM, Daszuta A (1999). Depletion in serotonin decreases neurogenesis in the dentate gyrus and the subventricular zone of adult rats. Neuroscience 89: 999–1002.
Caldarone BJ, Karthigeyan K, Harrist A, Hunsberger JG, Wittmack E, King SL et al (2003). Sex differences in response to oral amitriptyline in three animal models of depression in C57BL/6J mice. Psychopharmacology (Berl) 170: 94–101.
Cameron HA, McKay RD (2001). Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J Comp Neurol 435: 406–417.
Chaki S, Yoshikawa R, Hirota S, Shimazaki T, Maeda M, Kawashima N et al (2004). MGS0039: a potent and selective group II metabotropic glutamate receptor antagonist with antidepressant-like activity. Neuropharmacology 46: 457–467.
Conti AC, Cryan JF, Dalvi A, Lucki I, Blendy JA (2002). cAMP response element-binding protein is essential for the upregulation of brain-derived neurotrophic factor transcription but not the behavioral or endocrine responses to antidepressant drugs. J Neurosci 22: 3262–3268.
Crabbe JC, Wahlsten D, Dudek BC (1999). Genetics of mouse behavior: interactions with laboratory environment. Science 284: 1670–1672.
Crowley JJ, Blendy JA, Lucki I (2005). Strain-dependent antidepressant-like effects of citalopram in the mouse tail suspension test. Psychopharmacology (Berl) 183: 257–264.
Cryan JF, Mombereau C, Vassout A (2005). The tail suspension test as a model for assessing antidepressant activity: review of pharmacological and genetic studies in mice. Neurosci Biobehav Rev 29: 571–625.
Czeh B, Michaelis T, Watanabe T, Frahm J, de Biurrun G, van Kampen M et al (2001). Stress-induced changes in cerebral metabolites hippocampal volume and cell proliferation are prevented by antidepressant treatment with tianeptine. Proc Natl Acad Sci USA 98: 12796–12801.
De Foubert G, Carney SL, Robinson CS, Destexhe EJ, Tomlinson R, Hicks CA et al (2004). Fluoxetine-induced change in rat brain expression of brain-derived neurotrophic factor varies depending on length of treatment. Neuroscience 128: 597–604.
Dowlatshahi D, MacQueen GM, Wang JF, Young LT (1998). Increased temporal cortex CREB concentrations and antidepressant treatment in major depression. Lancet 352: 1754–1755.
Drigues N, Poltyrev T, Bejar C, Weinstock M, Youdim MB (2003). cDNA gene expression profile of rat hippocampus after chronic treatment with antidepressant drugs. J Neural Transm 110: 1413–1436.
Dulawa SC, Holick KA, Gundersen B, Hen R (2004). Effects of chronic fluoxetine in animal models of anxiety and depression. Neuropsychopharmacology 29: 1321–1330.
Duman RS, Heninger GR, Nestler EJ (1997). A molecular and cellular theory of depression. Arch Gen Psychiatry 54: 597–606.
Duman RS, Nakagawa S, Malberg J (2001). Regulation of adult neurogenesis by antidepressant treatment. Neuropsychopharmacology 25: 836–844.
Encinas JM, Vaahtokari A, Enikolopov G (2006). Fluoxetine targets early progenitor cells in the adult brain. Proc Natl Acad Sci USA 103: 8233–8238.
Gobbel GT, Bellinzona M, Vogt AR, Gupta N, Fike JR, Chan PH (1998). Response of postmitotic neurons to X-irradiation: implications for the role of DNA damage in neuronal apoptosis. J Neurosci 18: 147–155.
Gould E (1999). Serotonin and hippocampal neurogenesis. Neuropsychopharmacology 21 (2 Suppl): 46S–51S.
Guo Y, Weller P, Farrell E, Cheung P, Fitch B, Clark D et al (2006). In silico pharmacogenetics of warfarin metabolism. Nat Biotechnol 24: 531–536.
Hajszan T, MacLusky NJ, Leranth C (2005). Short-term treatment with the antidepressant fluoxetine triggers pyramidal dendritic spine synapse formation in rat hippocampus. Eur J Neurosci 21: 1299–1303.
Hastings NB, Gould E (1999). Rapid extension of axons into the CA3 region by adult-generated granule cells. J Comp Neurol 413: 146–154.
Hayes NL, Nowakowski RS (2002). Dynamics of cell proliferation in the adult dentate gyrus of two inbred strains of mice. Brain Res Dev Brain Res 134: 77–85.
Hillebrandt S, Wasmuth HE, Weiskirchen R, Hellerbrand C, Keppeler H, Werth A et al (2005). Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat Genet 37: 835–843.
Holmes A, Li Q, Koenig EA, Gold E, Stephenson D, Yang RJ et al (2005). Phenotypic assessment of galanin overexpressing and galanin receptor R1 knockout mice in the tail suspension test for depression-related behavior. Psychopharmacology (Berl) 178: 276–285.
Hoshaw BA, Malberg JE, Lucki I (2005). Central administration of IGF-I and BDNF leads to long-lasting antidepressant-like effects. Brain Res 1037: 204–208.
Kaplan MS, Hinds JW (1977). Neurogenesis in the adult rat: electron microscopic analysis of light radioautographs. Science 197: 1092–1094.
Katz MM, Tekell JL, Bowden CL, Brannan S, Houston JP, Berman N et al (2004). Onset and early behavioral effects of pharmacologically different antidepressants and placebo in depression. Neuropsychopharmacology 29: 566–579.
Kee N, Teixeira CM, Wang AH, Frankland PW (2007). Preferential incorporation of adult-generated granule cells into spatial memory networks in the dentate gyrus. Nat Neurosci 10: 355–362.
Kempermann G, Chesler EJ, Lu L, Williams RW, Gage FH (2006). Natural variation and genetic covariance in adult hippocampal neurogenesis. Proc Natl Acad Sci USA 103: 780–785.
Kempermann G, Gage FH (2002). Genetic influence on phenotypic differentiation in adult hippocampal neurogenesis. Brain Res Dev Brain Res 134: 1–12.
Kempermann G, Jessberger S, Steiner B, Kronenberg G (2004). Milestones of neuronal development in the adult hippocampus. Trends Neurosci 27: 447–452.
Kempermann G, Kuhn HG, Gage FH (1997). Genetic influence on neurogenesis in the dentate gyrus of adult mice. Proc Natl Acad Sci USA 94: 10409–10414.
Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR et al (2003). The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). JAMA 289: 3095–3105.
Khawaja X, Xu J, Liang JJ, Barrett JE (2004). Proteomic analysis of protein changes developing in rat hippocampus after chronic antidepressant treatment: Implications for depressive disorders and future therapies. J Neurosci Res 75: 451–460.
Koran LM, Cain JW, Dominguez RA, Rush AJ, Thiemann S (1996). Are fluoxetine plasma levels related to outcome in obsessive-compulsive disorder? Am J Psychiatry 153: 1450–1454.
Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y et al (2006). Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet 38: 888–895.
Liu X, Gershenfeld HK (2001). Genetic differences in the tail-suspension test and its relationship to imipramine response among 11 inbred strains of mice. Biol Psychiatry 49: 575–581.
Liu X, Gershenfeld HK (2003). An exploratory factor analysis of the Tail Suspension Test in 12 inbred strains of mice and an F2 intercross. Brain Res Bull 60: 223–231.
Lucki I, Dalvi A, Mayorga AJ (2001). Sensitivity to the effects of pharmacologically selective antidepressants in different strains of mice. Psychopharmacology (Berl) 155: 315–322.
Malagie I, Trillat AC, Jacquot C, Gardier AM (1995). Effects of acute fluoxetine on extracellular serotonin levels in the raphe: an in vivo microdialysis study. Eur J Pharmacol 286: 213–217.
Malberg JE, Duman RS (2003). Cell proliferation in adult hippocampus is decreased by inescapable stress: reversal by fluoxetine treatment. Neuropsychopharmacology 28: 1562–1571.
Malberg JE, Eisch AJ, Nestler EJ, Duman RS (2000). Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J Neurosci 20: 9104–9110.
McMahon FJ, Buervenich S, Charney D, Lipsky R, Rush AJ, Wilson AF et al (2006). Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. Am J Hum Genet 78: 804–814.
Mignone JL, Kukekov V, Chiang AS, Steindler D, Enikolopov G (2004). Neural stem and progenitor cells in nestin-GFP transgenic mice. J Comp Neurol 469: 311–324.
Minturn JE, Geschwind DH, Fryer HJ, Hockfield S (1995). Early postmitotic neurons transiently express TOAD-64 a neural specific protein. J Comp Neurol 355: 369–379.
Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM (2002). Neurobiology of depression. Neuron 34: 13–25.
Newman JC, Weiner AM (2005). L2L: a simple tool for discovering the hidden significance in microarray expression data. Genome Biol 6: R81.
Newton SS, Collier EF, Hunsberger J, Adams D, Terwilliger R, Selvanayagam E et al (2003). Gene profile of electroconvulsive seizures: induction of neurotrophic and angiogenic factors. J Neurosci 23: 10841–10851.
Nibuya M, Nestler EJ, Duman RS (1996). Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. J Neurosci 16: 2365–2372.
Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB et al (2004). Candidate genes pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry 9: 1007–1029.
Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E et al (2004). Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol 2: e393.
Porsolt RD, Bertin A, Jalfre M (1977). Behavioral despair in mice: a primary screening test for antidepressants. Arch Int Pharmacodyn Ther 229: 327–336.
Rola R, Raber J, Rizk A, Otsuka S, VandenBerg SR, Morhardt DR et al (2004). Radiation-induced impairment of hippocampal neurogenesis is associated with cognitive deficits in young mice. Exp Neurol 188: 316–330.
Ross SB, Renyi AL (1975). Tricyclic antidepressant agents. II. Effect of oral administration on the uptake of 3-H-noradrenaline and 14-C-5-hydroxytryptamine in slices of the midbrain-hypothalamus region of the rat. Acta Pharmacol Toxicol (Copenh) 36 (Suppl 5): 395–408.
Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D et al (2006). Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry 163: 1905–1917.
Saarelainen T, Hendolin P, Lucas G, Koponen E, Sairanen M, MacDonald E et al (2003). Activation of the TrkB neurotrophin receptor is induced by antidepressant drugs and is required for antidepressant-induced behavioral effects. J Neurosci 23: 349–357.
Sairanen M, Lucas G, Ernfors P, Castren M, Castren E (2005). Brain-derived neurotrophic factor and antidepressant drugs have different but coordinated effects on neuronal turnover proliferation and survival in the adult dentate gyrus. J Neurosci 25: 1089–1094.
Santarelli L, Saxe M, Gross C, Surget A, Battaglia F, Dulawa S et al (2003). Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science 301: 805–809.
Serretti A, Benedetti F, Zanardi R, Smeraldi E (2005). The influence of serotonin transporter promoter polymorphism (SERTPR) and other polymorphisms of the serotonin pathway on the efficacy of antidepressant treatments. Prog Neuropsychopharmacol Biol Psychiatry 29: 1074–1084.
Sheline YI, Gado MH, Kraemer HC (2003). Untreated depression and hippocampal volume loss. Am J Psychiatry 160: 1516–1518.
Sheline YI, Wang PW, Gado MH, Csernansky JG, Vannier MW (1996). Hippocampal atrophy in recurrent major depression. Proc Natl Acad Sci USA 93: 3908–3913.
Shirai K, Mizui T, Suzuki Y, Kobayashi Y, Nakano T, Shirao T (2006). Differential effects of X-irradiation on immature and mature hippocampal neurons in vitro. Neurosci Lett 399: 57–60.
Steiner B, Kronenberg G, Jessberger S, Brandt MD, Reuter K, Kempermann G (2004). Differential regulation of gliogenesis in the context of adult hippocampal neurogenesis in mice. Glia 46: 41–52.
Steru L, Chermat R, Thierry B, Simon P (1985). The tail suspension test: a new method for screening antidepressants in mice. Psychopharmacology (Berl) 85: 367–370.
Swanson CJ, Blackburn TP, Zhang X, Zheng K, Xu ZQ, Hokfelt T et al (2005). Anxiolytic- and antidepressant-like profiles of the galanin-3 receptor (Gal3) antagonists SNAP 37889 and SNAP 398299. Proc Natl Acad Sci USA 102: 17489–17494.
Toni N, Teng EM, Bushong EA, Aimone JB, Zhao C, Consiglio A et al (2007). Synapse formation on neurons born in the adult hippocampus. Nat Neurosci 10: 727–734.
Trullas R, Jackson B, Skolnick P (1989). Genetic differences in a tail suspension test for evaluating antidepressant activity. Psychopharmacology (Berl) 99: 287–288.
Videbech P, Avnkilde BR (2004). Hippocampal volume and depression: a meta-analysis of MRI studies. Am J Psychiatry 161: 1957–1966.
Wahlsten D, Metten P, Phillips TJ, Boehm II SL, Burkhart-Kasch S, Dorow J et al (2003). Different data from different labs: lessons from studies of gene–environment interaction. J Neurobiol 54: 283–311.
Acknowledgements
We thank Brandon Young and Bradley Long for invaluable work in producing the microarray data (Genomics Core, Scripps Florida), Paul Kenny (Scripps Florida), and Tim Wiltshire (Genomic Institute of the Novartis Research Foundation) for their thoughtful comments and suggestions, and Gerd Kempermann (Max Delbrück Center for Molecular Medicine) for consultation and advice regarding BrdU labeling. This study was supported by a grant from the state of Florida.
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclosure/Conflict of Interests
The authors do not have any conflict of interests to disclose.
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary information
Rights and permissions
About this article
Cite this article
Miller, B., Schultz, L., Gulati, A. et al. Genetic Regulation of Behavioral and Neuronal Responses to Fluoxetine. Neuropsychopharmacol 33, 1312–1322 (2008). https://doi.org/10.1038/sj.npp.1301497
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.npp.1301497
Keywords
This article is cited by
-
Obligatory roles of dopamine D1 receptors in the dentate gyrus in antidepressant actions of a selective serotonin reuptake inhibitor, fluoxetine
Molecular Psychiatry (2020)
-
Electroconvulsive therapy modulates plasma pigment epithelium-derived factor in depression: a proteomics study
Translational Psychiatry (2017)
-
Kdm6b and Pmepa1 as Targets of Bioelectrically and Behaviorally Induced Activin A Signaling
Molecular Neurobiology (2016)
-
Identifying individual differences of fluoxetine response in juvenile rhesus monkeys by metabolite profiling
Translational Psychiatry (2014)
-
Hippocampal HDAC4 Contributes to Postnatal Fluoxetine-Evoked Depression-Like Behavior
Neuropsychopharmacology (2014)