Abstract
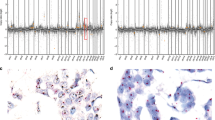
Five breast cancer subtypes have been described in sporadic breast cancer (SBC) using expression arrays: basal-like, ERBB2, normal breast-like, luminal A and B. These molecular subtypes show different genomic aberration patterns (GAPs). Recently, our group described these breast cancer subtypes in 50 non-BRCA1/2 familial tumors using immunohistochemistry assays. We extended this study to the other classes of familial breast cancer (FBC), including 62 tumors (18 BRCA1, 16 BRCA2 and 28 non-BRCA1/2), with the same panel of 25 immunohistochemical (IHC) markers and histological grade obtaining a similar classification. We combined these data with results generated by a 1 Mb BAC array-based CGH study to evaluate the genomic aberrations of each group. We found that BRCA1-related tumors are preferentially basal-like, whereas non-BRCA1/2 familial tumors are mainly luminal A subtype. We described distinct GAPs related to each IHC subtype. Basal tumors had a greater number of gains/losses, while luminal B tumors had more high-level DNA amplifications. Our data are similar to those obtained in SBC studies, highlighting the existence of distinct genetic pathways of tumor evolution, common to both SBC and FBC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Abbreviations
- aCGH:
-
array-based comparative genomic hybridization
- CNA:
-
copy number of genomic aberrations
- ER:
-
estrogen receptor
- FBC:
-
familial breast cancer
- GAP:
-
genomic aberration pattern
- IHC:
-
immunohistochemical
- SBC:
-
sporadic breast cancer
References
Adem C, Soderberg CL, Hafner K, Reynolds C, Slezak JM, Sinclair CS et al. (2004). ERBB2, TBX2, RPS6KB1, and MYC alterations in breast tissues of BRCA1 and BRCA2 mutation carriers. Genes Chromosomes Cancer 41: 1–11.
Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H et al. (2004). Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res 64: 8534–8540.
Alvarez S, Diaz-Uriarte R, Osorio A, Barroso A, Melchor L, Paz MF et al. (2005). A predictor based on the somatic genomic changes of the BRCA1/BRCA2 breast cancer tumors identifies the non-BRCA1/BRCA2 tumors with BRCA1 promoter hypermethylation. Clin Cancer Res 11: 1146–1153.
Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE et al. (2006). Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer 45: 1033–1040.
Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K et al. (2006). Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295: 2492–2502.
Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL et al. (2006). Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541.
Diez O, Osorio A, Duran M, Martinez-Ferrandis JI, de la Hoya M, Salazar R et al. (2003). Analysis of BRCA1 and BRCA2 genes in Spanish breast/ovarian cancer patients: a high proportion of mutations unique to Spain and evidence of founder effects. Hum Mutat 22: 301–312.
Elsheikh S, Green AR, Aleskandarany MA, Grainge M, Paish CE, Lambros MB et al. (2007). CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res Treat; e-pub ahead of print.
Greshock J, Naylor TL, Margolin A, Diskin S, Cleaver SH, Futreal PA et al. (2004). 1-Mb resolution array-based comparative genomic hybridization using a BAC clone set optimized for cancer gene analysis. Genome Res 14: 179–187.
Grushko TA, Blackwood MA, Schumm PL, Hagos FG, Adeyanju MO, Feldman MD et al. (2002). Molecular-cytogenetic analysis of HER-2/neu gene in BRCA1-associated breast cancers. Cancer Res 62: 1481–1488.
Herrero J, Al-Shahrour F, Diaz-Uriarte R, Mateos A, Vaquerizas JM, Santoyo J et al. (2003). GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res 31: 3461–3467.
Herrero J, Valencia A, Dopazo J . (2001). A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics 17: 126–136.
Honrado E, Benitez J, Palacios J . (2005a). The molecular pathology of hereditary breast cancer: genetic testing and therapeutic implications. Mod Pathol 18: 1305–1320.
Honrado E, Osorio A, Milne RL, Paz MF, Melchor L, Cascon A et al. (2007). Immunohistochemical classification of non-BRCA1/2 tumors identifies different groups that demonstrate the heterogeneity of BRCAX families. Mod Pathol 20: 1298–1306.
Honrado E, Osorio A, Palacios J, Milne RL, Sanchez L, Diez O et al. (2005b). Immunohistochemical expression of DNA repair proteins in familial breast cancer differentiate BRCA2-associated tumors. J Clin Oncol 23: 7503–7511.
Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF et al. (2006). The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7: 96.
Jonsson G, Naylor TL, Vallon-Christersson J, Staaf J, Huang J, Ward MR et al. (2005). Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res 65: 7612–7621.
Lacroix M, Leclercq G . (2005). The ‘portrait’ of hereditary breast cancer. Breast Cancer Res Treat 89: 297–304.
Lakhani SR, Jacquemier J, Sloane JP, Gusterson BA, Anderson TJ, van de Vijver MJ et al. (1998). Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst 90: 1138–1145.
Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L et al. (2002). The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol 20: 2310–2318.
Melchor L, Honrado E, Huang J, Alvarez S, Naylor TL, Garcia MJ et al. (in press). Estrogen receptor status could modulate the genomic pattern in familial and sporadic breast cancer: a study based on array CGH. Clin Cancer Res (in press).
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S et al. (1994). A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266: 66–71.
Oldenburg RA, Kroeze-Jansema K, Meijers-Heijboer H, van Asperen CJ, Hoogerbrugge N, van Leeuwen I et al. (2006). Characterization of familial non-BRCA1/2 breast tumors by loss of heterozygosity and immunophenotyping. Clin Cancer Res 12: 1693–1700.
Osorio A, Barroso A, Martinez B, Cebrian A, San Roman JM, Lobo F et al. (2000). Molecular analysis of the BRCA1 and BRCA2 genes in 32 breast and/or ovarian cancer Spanish families. BJC 82: 1266–1270.
Palacios J, Honrado E, Osorio A, Cazorla A, Sarrio D, Barroso A et al. (2003). Immunohistochemical characteristics defined by tissue microarray of hereditary breast cancer not attributable to BRCA1 or BRCA2 mutations: differences from breast carcinomas arising in BRCA1 and BRCA2 mutation carriers. Clin Cancer Res 9: 3606–3614.
Palacios J, Honrado E, Osorio A, Cazorla A, Sarrio D, Barroso A et al. (2005). Phenotypic characterization of BRCA1 and BRCA2 tumors based in a tissue microarray study with 37 immunohistochemical markers. Breast Cancer Res Treat 90: 5–14.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al. (2000). Molecular portraits of human breast tumours. Nature 406: 747–752.
Reis-Filho JS, Savage K, Lambros MB, James M, Steele D, Jones RL et al. (2006). Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: an immunohistochemical and chromogenic in situ hybridisation analysis. Mod Pathol 19: 999–1009.
Rodriguez-Pinilla SM, Jones RL, Lambros MB, Arriola E, Savage K, James M et al. (2007). MYC amplification in breast cancer: a chromogenic in situ hybridisation study. J Clin Pathol 60: 1017–1023.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H et al. (2001). Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874.
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A et al. (2003). Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100: 8418–8423.
Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A et al. (2003). Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA 100: 10393–10398.
Staff S, Isola J, Tanner M . (2003). Haplo-insufficiency of BRCA1 in sporadic breast cancer. Cancer Res 63: 4978–4983.
Tirkkonen M, Johannsson O, Agnarsson BA, Olsson H, Ingvarsson S, Karhu R et al. (1997). Distinct somatic genetic changes associated with tumor progression in carriers of BRCA1 and BRCA2 germ-line mutations. Cancer Res 57: 1222–1227.
Tischkowitz MD, Foulkes WD . (2006). The basal phenotype of BRCA1-related breast cancer: past, present and future. Cell Cycle 5: 963–967.
Turner NC, Reis-Filho JS . (2006). Basal-like breast cancer and the BRCA1 phenotype. Oncogene 25: 5846–5853.
Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D et al. (2006). BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 26: 2126–2132.
van Beers EH, van Welsem T, Wessels LF, Li Y, Oldenburg RA, Devilee P et al. (2005). Comparative genomic hybridization profiles in human BRCA1 and BRCA2 breast tumors highlight differential sets of genomic aberrations. Cancer Res 65: 822–827.
Vaquerizas JM, Conde L, Yankilevich P, Cabezon A, Minguez P, Diaz-Uriarte R et al. (2005). GEPAS, an experiment-oriented pipeline for the analysis of microarray gene expression data. Nucleic Acids Res 33: W616–W620.
Vaquerizas JM, Dopazo J, Diaz-Uriarte R . (2004). DNMAD: web-based diagnosis and normalization for microarray data. Bioinformatics 20: 3656–3658.
Vincent-Salomon A, Gruel N, Lucchesi C, Mac Grogan G, Dendale R, Sigal-Zafrani B et al. (2007). Identification of typical medullary breast carcinoma as a genomic sub-group of basal-like carcinomas, a heterogeneous new molecular entity. Breast Cancer Res 9: R24.
Wei M, Grushko TA, Dignam J, Hagos F, Nanda R, Sveen L et al. (2005). BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res 65: 10692–10699.
Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J et al. (1995). Identification of the breast cancer susceptibility gene BRCA2. Nature 378: 789–792.
Yehiely F, Moyano JV, Evans JR, Nielsen TO, Cryns VL . (2006). Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol Med 12: 537–544.
Acknowledgements
We thank the Spanish National Tumor Bank Network, and Luz Álvarez and Miguel Urioste from the Familial Cancer Unit for the collection of the samples. For their technical assistance, we thank the Immunohistological Unit, and Carmen Martin and Juan C Cigudosa from the Molecular Cytogenetics Group, at the Spanish National Cancer Center and C Angelica Medina from Medical Genetics Division of the University of Pennsylvania. We also thank Susanna Leskelä for English assistance. LM Grant sponsor: Spanish Ministry of Education and Science FPU AP-2004-0448. Short stays sponsorships: ICRETT/05/063 and EMBO Short-Term Fellowship ASTF 162-05. MJG was supported by the Fundación Científica de la Asociación Española contra el Cáncer. This study has been partially funded by projects SAF03-02497 and SAF 06-06149 from the Spanish Ministry of Education and Science; and by a grant from Breast Cancer Research Foundation (BCRF) to KLN.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Rights and permissions
About this article
Cite this article
Melchor, L., Honrado, E., García, M. et al. Distinct genomic aberration patterns are found in familial breast cancer associated with different immunohistochemical subtypes. Oncogene 27, 3165–3175 (2008). https://doi.org/10.1038/sj.onc.1210975
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1210975
Keywords
This article is cited by
-
RANK-c attenuates aggressive properties of ER-negative breast cancer by inhibiting NF-κB activation and EGFR signaling
Oncogene (2018)
-
RNA profiling reveals familial aggregation of molecular subtypes in non-BRCA1/2 breast cancer families
BMC Medical Genomics (2014)
-
Should BRCA2 mutation carriers avoid neoadjuvant chemotherapy?
Medical Oncology (2014)
-
Short telomeres are frequent in hereditary breast tumors and are associated with high tumor grade
Breast Cancer Research and Treatment (2013)
-
DNA copy number profiling reveals extensive genomic loss in hereditary BRCA1 and BRCA2 ovarian carcinomas
British Journal of Cancer (2013)