Abstract
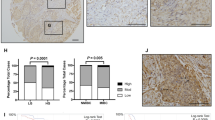
Inter-α-trypsin inhibitors (ITIs) are protease inhibitors stabilizing the extracellular matrix. ITIs consist of one light (bikunin) and two heavy chains (ITIHs). We have recently characterized ITIH5, a novel member of the ITIH gene family, and showed that its messenger RNA is lost in a high proportion of breast tumours. In the present study, an ITIH5-specific polyclonal antibody was generated, validated with western blot and used for immunohistochemical analysis on a tissue microarray; ITIH5 was strongly expressed in epithelial cells of normal breast (n=11/15), while it was lost or strongly reduced in 42% (92/217) of invasive breast cancers. ITIH5 expression in invasive carcinomas was associated with positive expression of oestrogen receptor (P=0.008) and histological grade (P=0.024). Correlation of ITIH5 expression with clinical outcome revealed that patients with primary tumours retaining abundant ITIH5 expression had longer recurrence-free survival (RFS; P=0.037) and overall survival (OS; P=0.044), compared to those with reduced expression (mean RFS: 102 vs 78 months; mean OS: 120 vs 105 months). Methylation-specific PCR analysis frequently showed strong methylation of the ITIH5 promoter in primary breast tumours (41%, n=109) and breast cancer cell lines (n=6). Methylation was significantly associated with mRNA loss (P<0.001; n=39), and ITIH5 expression was induced after treatment of tumour cell lines with the demethylating agent 5-aza-2′-deoxycytidine. Moreover, ITIH5 promoter methylation was significantly associated with reduced OS (P=0.008). The cellular function of ITIH5 was evaluated by forced expression of a full-length ITIH5 complementary DNA in the breast cancer cell line MDA-MB-231, which does not endogenously express ITIH5. ITIH5-expressing clones showed a 40% reduced proliferation rate compared to mock-transfected cells. Overall, these data show that promoter methylation-mediated loss of ITIH5 expression is associated with unfavourable outcome in breast cancer patients, and thus ITIH5 could be used as a prognostic marker, although this marker is not multivariate independent due to its close association with ER expression. Our data indicate that ITIH5 is a candidate class II tumour suppressor gene and could be involved in tumour progression, invasion and metastasis, as its absence is associated with increased proliferation rates and a prognostic value indicating poor clinical outcome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Abbreviations
- CI:
-
confidence interval
- DAC:
-
5-aza-2′-deoxycytidine
- DCIS:
-
ductal carcinoma in situ
- ER:
-
oestrogen receptor
- IRS:
-
immunoreactivity score
- ITI:
-
inter-α-trypsin inhibitor
- ITIH:
-
inter-α-trypsin inhibitor heavy chain
- MSP:
-
methylation-specific PCR
- OS:
-
overall survival
- PR:
-
progesterone receptor
- RFS:
-
recurrence-free survival
- TMA:
-
tissue microarray
- TSA:
-
trichostatin A
References
Atmani F, Khan SR . (1999). Role of inter-alpha-inhibitor and its related proteins in urolithiasis. Purification of an inter-alpha-inhibitor related protein from the bovine kidney. Urol Res 27: 57–61.
Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK et al. (1999). Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res 59: 798–802.
Balduyck M, Albani D, Jourdain M, Mizon C, Tournoys A, Drobecq H et al. (2000). Inflammation-induced systemic proteolysis of inter-alpha-inhibitor in plasma from patients with sepsis. J Lab Clin Med 135: 188–198.
Bost F, Diarra-Mehrpour M, Martin JP . (1998). Inter-alpha-trypsin inhibitor proteoglycan family—a group of proteins binding and stabilizing the extracellular matrix. Eur J Biochem 252: 339–346.
Bourguignon J, Borghi H, Sesboue R, Diarra-Mehrpour M, Bernaudin JF, Metayer J et al. (1999). Immunohistochemical distribution of inter-alpha-trypsin inhibitor chains in normal and malignant human lung tissue. J Histochem Cytochem 47: 1625–1632.
Chen L, Mao SJ, McLean LR, Powers RW, Larsen WJ . (1994). Proteins of the inter-alpha-trypsin inhibitor family stabilize the cumulus extracellular matrix through their direct binding with hyaluronic acid. J Biol Chem 269: 28282–28287.
Dahl E, Kristiansen G, Gottlob K, Klaman I, Ebner E, Hinzmann B et al. (2006). Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin Cancer Res 12: 3950–3960.
Dahl E, Sadr-Nabavi A, Klopocki E, Betz B, Grube S, Kreutzfeld R et al. (2005). Systematic identification and molecular characterization of genes differentially expressed in breast and ovarian cancer. J Pathol 205: 21–28.
Das PM, Singal R . (2004). DNA methylation and cancer. J Clin Oncol 22: 4632–4642.
Diarra-Mehrpour M, Bourguignon J, Sesboue R, Mattei MG, Passage E, Salier JP et al. (1989). Human plasma inter-alpha-trypsin inhibitor is encoded by four genes on three chromosomes. Eur J Biochem 179: 147–154.
Elston CW, Ellis IO . (1991). Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19: 403–410.
Enghild JJ, Salvesen G, Hefta SA, Thogersen IB, Rutherfurd S, Pizzo SV . (1991). Chondroitin 4-sulfate covalently cross-links the chains of the human blood protein pre-alpha-inhibitor. J Biol Chem 266: 747–751.
Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG . (1999). Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res 59: 67–70.
Esteller M . (2005). DNA methylation and cancer therapy: new developments and expectations. Curr Opin Oncol 17: 55–60.
Fujita Y, Ezura Y, Emi M, Sato K, Takada D, Iino Y et al. (2004). Hypercholesterolemia associated with splice-junction variation of inter-alpha-trypsin inhibitor heavy chain 4 (ITIH4) gene. J Hum Genet 49: 24–28.
Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF et al. (1995). E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res 55: 5195–5199.
Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB . (1996). Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93: 9821–9826.
Himmelfarb M, Klopocki E, Grube S, Staub E, Klaman I, Hinzmann B et al. (2004). ITIH5, a novel member of the inter-α-trypsin inhibitor heavy chain family is downregulated in breast cancer. Cancer Lett 204: 69–77.
Huang L, Yoneda M, Kimata K . (1993). A serum-derived hyaluronan-associated protein (SHAP) is the heavy chain of the inter alpha-trypsin inhibitor. J Biol Chem 268: 26725–26730.
Janssen U, Thomas G, Glant T, Phillips A . (2001). Expression of inter-alpha-trypsin inhibitor and tumor necrosis factor-stimulated gene 6 in renal proximal tubular epithelial cells. Kidney Int 60: 126–136.
Jessen TE, Odum L . (2003). Role of tumour necrosis factor stimulated gene 6 (TSG-6) in the coupling of inter-alpha-trypsin inhibitor to hyaluronan in human follicular fluid. Reproduction 125: 27–31.
Kobayashi H, Gotoh J, Hirashima Y, Fujie M, Sugino D, Terao T . (1995). Inhibitory effect of a conjugate between human urokinase and urinary trypsin inhibitor on tumor cell invasion in vitro. J Biol Chem 270: 8361–8366.
Onda H, Kasuya H, Takakura K, Hori T, Imaizumi T, Takeuchi T et al. (1999). Identification of genes differentially expressed in canine vasospastic cerebral arteries after subarachnoid hemorrhage. J Cereb Blood Flow Metab 19: 1279–1288.
Paris S, Sesboue R, Delpech B, Chauzy C, Thiberville L, Martin JP et al. (2002). Inhibition of tumor growth and metastatic spreading by overexpression of inter-alpha-trypsin inhibitor family chains. Int J Cancer 97: 615–620.
Pineiro M, Andres M, Iturralde M, Carmona S, Hirvonen J, Pyorala S et al. (2004). ITIH4 (inter-alpha-trypsin inhibitor heavy chain 4) is a new acute-phase protein isolated from cattle during experimental infection. Infect Immun 72: 3777–3782.
Remmele W, Stegner HE . (1987). Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 8: 138–140.
Rugg MS, Willis AC, Mukhopadhyay D, Hascall VC, Fries E, Fulop C et al. (2005). Characterization of complexes formed between TSG-6 and inter-alpha-inhibitor that act as intermediates in the covalent transfer of heavy chains onto hyaluronan. J Biol Chem 280: 25674–25686.
Sager R . (1989). Tumor suppressor genes: the puzzle and the promise. Science 246: 1406–1412.
Sanggaard KW, Karring H, Valnickova Z, Thogersen IB, Enghild JJ . (2005). The TSG-6 and I alpha I interaction promotes a transesterification cleaving the protein-glycosaminoglycan-protein (PGP) cross-link. J Biol Chem 280: 11936–11942.
Shao G, Berenguer J, Borczuk AC, Powell CA, Hei TK, Zhao Y . (2006). Epigenetic inactivation of Betaig-h3 gene in human cancer cells. Cancer Res 66: 4566–4573.
Sobin LH, Wittekind CH (eds) (2002). UICC: TNM Classification of Malignant Tumours 6th edn. New York: Wiley-Liss.
Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP et al. (2002). A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet 31: 141–149.
Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B et al. (2006). Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene 25: 3479–3488.
Yang X, Yan L, Davidson NE . (2001). DNA methylation in breast cancer. Endocr Relat Cancer 8: 115–127.
Yingsung W, Zhuo L, Morgelin M, Yoneda M, Kida D, Watanabe H et al. (2003). Molecular heterogeneity of the SHAP-hyaluronan complex. Isolation and characterization of the complex in synovial fluid from patients with rheumatoid arthritis. J Biol Chem 278: 32710–32718.
Yoneda M, Suzuki S, Kimata K . (1990). Hyaluronic acid associated with the surfaces of cultured fibroblasts is linked to a serum-derived 85-kDa protein. J Biol Chem 265: 5247–5257.
Zhang Z, Bast Jr RC, Yu Y, Li J, Sokoll LJ, Rai AJ et al. (2004). Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res 64: 5882–5890.
Zhao M, Yoneda M, Ohashi Y, Kurono S, Iwata H, Ohnuki Y et al. (1995). Evidence for the covalent binding of SHAP, heavy chains of inter-alpha-trypsin inhibitor, to hyaluronan. J Biol Chem 270: 26657–26663.
Zhuo L, Yoneda M, Zhao M, Yingsung W, Yoshida N, Kitagawa Y et al. (2001). Defect in SHAP-hyaluronan complex causes severe female infertility. A study by inactivation of the bikunin gene in mice. J Biol Chem 276: 7693–7696.
Acknowledgements
We thank Sonja von Serényi, Sevim Alkaya and Inge Losen for excellent technical assistance and Monika Klinkhammer-Schalke and Armin Pauer from the Tumor Registry Regensburg for continuous help in obtaining clinical follow-up data. This work is a research project within the German Human Genome Project and has been supported by the BMBF Grants 01KW0401 to ED and a grant from the RWTH Aachen (START program project ITIH5).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Rights and permissions
About this article
Cite this article
Veeck, J., Chorovicer, M., Naami, A. et al. The extracellular matrix protein ITIH5 is a novel prognostic marker in invasive node-negative breast cancer and its aberrant expression is caused by promoter hypermethylation. Oncogene 27, 865–876 (2008). https://doi.org/10.1038/sj.onc.1210669
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1210669
Keywords
This article is cited by
-
An Overview on the Emerging Role of the Plasma Protease Inhibitor Protein ITIH5 as a Metastasis Suppressor
Cell Biochemistry and Biophysics (2024)
-
Switched alternative splicing events as attractive features in lung squamous cell carcinoma
Cancer Cell International (2022)
-
Suppression of pancreatic cancer liver metastasis by secretion-deficient ITIH5
British Journal of Cancer (2021)
-
ITIH5, a p53-responsive gene, inhibits the growth and metastasis of melanoma cells by downregulating the transcriptional activity of KLF4
Cell Death & Disease (2021)
-
Prognostic DNA methylation markers for hormone receptor breast cancer: a systematic review
Breast Cancer Research (2020)