Abstract
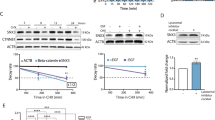
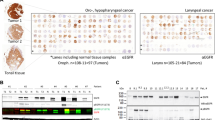
Carcinomas are tumors of epithelial origin accounting for over 80% of all human malignancies. A substantial body of evidence implicates oncogenic signaling by receptor tyrosine kinases (RTKs) in carcinoma development. Here we investigated the expression of Sef, a novel inhibitor of RTK signaling, in normal human epithelial tissues and derived malignancies. Human Sef (hSef) was highly expressed in normal epithelial cells of breast, prostate, thyroid gland and the ovarian surface. By comparison, substantial downregulation of hSef expression was observed in the majority of tumors originating from these epithelia. Among 186 primary carcinomas surveyed by RNA in situ hybridization, hSef expression was undetectable in 116 cases including 72/99 (73%) breast, 11/16 (69%) thyroid, 16/31 (52%) prostate and 17/40 (43%) ovarian carcinomas. Moderate reduction of expression was observed in 17/186, and marked reduction in 40/186 tumors. Only 13/186 cases including 12 low-grade and one intermediate grade tumor retained high hSef expression. The association of hSef downregulation and tumor progression was statistically significant (P<0.001). Functionally, ectopic expression of hSef suppressed proliferation of breast carcinoma cells, whereas inhibition of endogenous hSef expression accelerated fibroblast growth factor and epidermal growth factor-dependent proliferation of cervical carcinoma cells. The inhibitory effect of hSef on cell proliferation combined with consistent downregulation in human carcinoma indicates a tumor suppressor-like role for hSef, and implicates loss of hSef expression as a common mechanism in epithelial neoplasia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Abbreviations
- EOC:
-
epithelial ovarian cancers
- GG:
-
Gleason grade
- OSE:
-
ovarian surface epithelium
- RTK:
-
receptor tyrosine kinase
- Sef:
-
similar expression to FGF genes
References
Bell DA . (2005). Origins and molecular pathology of ovarian cancer. Mod Patho 18: S19–S32.
Blume-Jensen P, Hunter T . (2001). Oncogenic kinase signalling. Nature 411: 355–365.
Darby S, Sahadevan K, Khan MM, Robson CN, Leung HY, Gnanapragasam VJ . (2006). Loss of Sef (similar expression to FGF) expression is associated with high grade and metastatic prostate cancer. Oncogene 25: 4122–4127.
De Marzo AM, Coffey DS, Nelson WG . (1999). New concepts in tissue specificity for prostate cancer and BPH. Urology 53: 29–39.
Furthauer M, Lin W, Ang SL, Thisse B, Thisse C . (2002). Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol 4: 170–174.
Gimm O . (2001). Thyroid Cancer. Cancer Lett 163: 143–156.
Grose R, Dickson C . (2005). Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev 16: 179–186.
Harduf H, Halperin E, Reshef R, Ron D . (2005). Sef is synexpressed with FGFs during chick embryogenesis and its expression is differentially regulated by FGFs in the developing limb. Dev Dyn 233: 301–312.
Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C et al. (2006). Cancer statistics, 2006. CA Cancer J Clin 56: 106–130.
Kosary CL . (1994). FIGO stage, histology, histologic grade, age and race as prognostic factors in determining survival for cancers of the female gynecological system: an analysis of 1973-87 SEER cases of cancers of the endometrium, cervix, ovary, vulva, and vagina. Semin Surg Oncol 10: 31–46.
Matsumoto S, Kasumi F, Sakamoto G, Onda M, Nakamura Y, Emi M . (1997). Detailed deletion mapping of chromosome arm 3p in breast cancers: a 2-cM region on 3p14.3-21.1 and a 5-cM region on 3p24.3-25.1 commonly deleted in tumors. Genes Chromosomes Cancer 20: 268–274.
Normanno N, Bianco C, Strizzi L, Mancino M, Maiello MR, De Luca A et al. (2005). The ErbB receptors and their ligands in cancer: an overview. Curr Drug Targets 6: 243–257.
Preger E, Ziv I, Shabtay A, Sher I, Tsang M, Dawid IB et al. (2004). Alternative splicing generates an isoform of the human Sef gene with altered subcellular localization and specificity. Proc Natl Acad Sci USA 101: 1229–1234.
Shaoul E, Reich-Slotky R, Berman B, Ron D . (1995). Fibroblast growth factor receptors display both common and distinct signaling pathways. Oncogene 10: 1553–1561.
Sher I, Zisman-Rozen S, Eliahu L, Whitelock JM, Maas-Szabowski N, Yamada Y et al. (2006). Targeting perlecan in human keratinocytes reveals novel roles for perlecan in epidermal formation. J Biol Chem 281: 5178–5187.
Tavassoli FA, Devilee P . (2003). Genetics of Tumors of the Breast, and Female Genital Organs. IARC Press, pp 18.
Thompson N, Lyons J . (2005). Recent progress in targeting the Raf/MEK/ERK pathway with inhibitors in cancer drug discovery. Curr Opin Pharmacol 5: 350–356.
Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E . (2004). Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell 7: 33–44.
Tsang M, Friesel R, Kudoh T, Dawid IB . (2002). Identification of Sef, a novel modulator of FGF signalling. Nat Cell Biol 4: 165–169.
Xiong S, Zhao Q, Rong Z, Huang G, Huang Y, Chen P et al. (2003). hSef Inhibits PC-12 cell differentiation by interfering with Ras-mitogen-activated protein kinase MAPK signaling. J Biol Chem 278: 50273–50282.
Yang RB, Ng CK, Wasserman SM, Komuves LG, Gerritsen ME, Topper JN . (2003). A novel IL-17 receptor-like protein identified in human umbilical vein endothelial cells antagonizes basic fibroblast growth factor-induced signaling. J Biol Chem 278: 33232–33238.
Ziv I, Fuchs Y, Preger E, Shabtay A, Zilpa T, Dym N et al. (2006). Human Sef-a isoform utilizes different mechanisms to regulate RTK signaling pathways and subsequent cell fate. J Biol Chem 281: 39225–39235.
Acknowledgements
The authors thank Sharon Lubinsky-Mink and Dr Orit Goldsmidt for excellent technical assistance. This work was supported by grants from the ICRF (# 2004973), and the Israel Ministry of Health (# 1005314) to Dina Ron.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Supplementary information
Rights and permissions
About this article
Cite this article
Zisman-Rozen, S., Fink, D., Ben-Izhak, O. et al. Downregulation of Sef, an inhibitor of receptor tyrosine kinase signaling, is common to a variety of human carcinomas. Oncogene 26, 6093–6098 (2007). https://doi.org/10.1038/sj.onc.1210424
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1210424
Keywords
This article is cited by
-
Thymoquinone upregulates IL17RD in controlling the growth and metastasis of triple negative breast cancer cells in vitro
BMC Cancer (2022)
-
Loss of interleukin-17 receptor D promotes chronic inflammation-associated tumorigenesis
Oncogene (2021)
-
Delivery of the gene encoding the tumor suppressor Sef into prostate tumors by therapeutic-ultrasound inhibits both tumor angiogenesis and growth
Scientific Reports (2017)
-
A knowledgebase resource for interleukin-17 family mediated signaling
Journal of Cell Communication and Signaling (2015)
-
Characterization of Golgi scaffold proteins and their roles in compartmentalizing cell signaling
Journal of Molecular Histology (2014)