Abstract
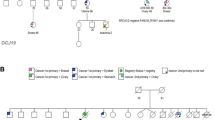
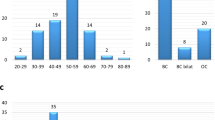
BRCA2 (BReast CAncer susceptibility gene 2) germline mutation carriers are at increased risk for breast and ovarian cancers. Mutations occurring in the ovarian cancer cluster region (OCCR) are linked to higher ovarian cancer and/or lower breast cancer risk(s) than mutations occurring elsewhere in BRCA2. Most BRCA2 germline mutations introduce premature termination codons (PTCs), making their mRNAs likely targets of nonsense-mediated mRNA decay (NMD), a mechanism that eliminates PTC-bearing transcripts to prevent expression of truncated proteins. Contradictory evidence exists regarding whether NMD can be triggered by PTCs located far upstream of the nearest exon–exon junction (EEJ). Since the OCCR comprises a major portion of the 4.9 kb exon 11 of BRCA2, we investigated if transcripts bearing PTCs in this large exon are unable to trigger NMD, and if this might contribute to the phenotypic difference associated with the OCCR. We examined cDNA from 18 carriers of PTC-introducing germline mutations located throughout BRCA2, and found that PTC-bearing transcripts were 1.4–3.3-fold less prevalent than their nonmutated counterparts irregardless of PTC position. We conclude that NMD can recognize PTCs up to 4.5 kb upstream of the nearest EEJ, demonstrating that a general inability of NMD to recognize PTCs in exon 11 is unlikely to explain the genotype–phenotype correlation associated with the OCCR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Chatr-Aryamontri A, Angelini M, Garelli E, Tchernia G, Ramenghi U, Dianzani I et al. (2004). Hum Mutat 24: 526–533.
Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH . (1998). Proc Natl Acad Sci USA 95: 5287–5292.
Cowles CR, Hirschhorn JN, Altshuler D, Lander ES . (2002). Nat Genet 32: 432–437.
Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR et al. (2001). Mol Cell 7: 273–282.
Gayther SA, Mangion J, Russell P, Seal S, Barfoot R, Ponder BA et al. (1997). Nat Genet 15: 103–105.
Gismondi V, Stagnaro P, Pedemonte S, Biticchi R, Presciuttini S, Grammatico P et al. (1998). Genes Chromosomes Cancer 22: 278–286.
Harries LW, Hattersley AT, Ellard S . (2004). Diabetes 53: 500–504.
Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE . (2004). Nat Genet 36: 801–808.
Lubinski J, Phelan CM, Ghadirian P, Lynch HT, Garber J, Weber B et al. (2004). Fam Cancer 3: 1–10.
Maquat LE . (2004). Nat Rev Mol Cell Biol 5: 89–99.
Maquat LE, Li X . (2001). RNA 7: 445–456.
Mizuta R, LaSalle JM, Cheng HL, Shinohara A, Ogawa H, Copeland N et al. (1997). Proc Natl Acad Sci USA 94: 6927–6932.
Neu-Yilik G, Gehring NH, Thermann R, Frede U, Hentze MW, Kulozik AE . (2001). EMBO J 20: 532–540.
Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S . (2002). Hum Mol Genet 11: 2805–2814.
Rodningen OK, Tonstad S, Saugstad OD, Ose L, Leren TP . (1999). Hum Mutat 13: 186–196.
Thompson D, Easton D . (2001). Am J Hum Genet 68: 410–419.
Thompson D, Easton D . (2004). J Mammary Gland Biol Neoplasia 9: 221–236.
Tournier I, Raux G, Di Fiore F, Marechal I, Leclerc C, Martin C et al. (2004). Hum Mutat 23: 379–384.
Venkitaraman AR . (2002). Cell 108: 171–182.
Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL . (1997). J Biol Chem 272: 31941–31944.
Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW . (2002). Science 297: 1143.
Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH et al. (2002). Science 297: 1837–1848.
Yang H, Li Q, Fan J, Holloman WK, Pavletich NP . (2005). Nature 433: 653–657.
Yoshida K, Miki Y . (2004). Cancer Sci 95: 866–871.
Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE . (1998). Mol Cell Biol 18: 5272–5283.
Acknowledgements
We thank Colette Bonnardel for her expert family data management, Nathalie Foray and Susanne Pauly for their excellent technical assistance, the Genethon (France) for the establishment of lymphoblastoid cell lines and HT Lynch for his long-term collaboration in this study. This work was supported by la Ligue Nationale Contre le Cancer – Comité du Rhône and the Interdisciplinary Health Research International Team on Breast Cancer Susceptibility (INHERIT BRCA) Research Program. MDW was funded by the Ministère de la Recherche and l'Association pour la Recherche sur le Cancer.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Oncogene website (http://www.nature.com/onc)
Supplementary information
Rights and permissions
About this article
Cite this article
Ware, M., DeSilva, D., Sinilnikova, O. et al. Does nonsense-mediated mRNA decay explain the ovarian cancer cluster region of the BRCA2 gene?. Oncogene 25, 323–328 (2006). https://doi.org/10.1038/sj.onc.1209033
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1209033
Keywords
This article is cited by
-
Nonsense-Mediated mRNA Decay: Mechanistic Insights and Physiological Significance
Molecular Biotechnology (2023)
-
Nonsense-mediated RNA decay: an emerging modulator of malignancy
Nature Reviews Cancer (2022)
-
Nonsense-mediated RNA decay and its bipolar function in cancer
Molecular Cancer (2021)
-
Mutations of BRCA2 in canine mammary tumors and their targeting potential in clinical therapy
BMC Veterinary Research (2020)
-
Quality and quantity control of gene expression by nonsense-mediated mRNA decay
Nature Reviews Molecular Cell Biology (2019)