Abstract
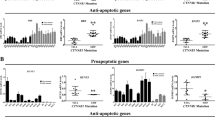
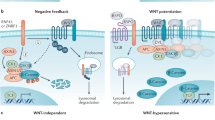
β-catenin is a multifunctional protein involved in both cadherin-mediated adhesion and the wnt signaling cascade. Mutations in exon 3 of β-catenin have been identified in many cancers. In addition to disruption of key serine and threonine residues, mutations are frequently reported in other residues in exon 3 that are not kinase substrates. The most frequently mutated nonserine/threonine residues are D32 and G34. Since D32 and G34 are part of the ubiquitination destruction motif, DSGΦXS, we hypothesize that this motif may contribute to disruption of β-catenin homeostasis and lead to cellular transformation. We demonstrate that the mutants D32A and G34A exhibit no change in phosphorylation by GSK3β, but display reduced ubiquitination compared to wild-type and S33A mutant β-catenin. To assess the functional implications of these mutations, we created stable MDCK cell lines expressing these constructs. We found that stable cell lines harboring D32A-mutated β-catenin were highly transformed, while S33A and G34 demonstrated only weak transforming properties in our assays. Despite altered ubiquitination status and increased transformation, the D32A mutant cell line does not display transcriptional activation of standard target genes. Therefore, D32A mutation may mediate transformation by an alternative β-catenin-mediated signaling pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Aberle H, Bauer A, Stappert J, Kispert A and Kemler R . (1997). EMBO J., 16, 3797–3804.
Al-Fageeh M, Li Q, Mohaiza Dashwood W, Myzak MC and Dashwood RH . (2004). Oncogene, 23, 4839–4846.
Besnard-Guerin C, Belaidouni N, Lassot I, Segeral E, Jobart A, Marchal C and Benarous R . (2004). J. Biol. Chem., 279, 788–795.
Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, Margottin-Goguet F, Jackson PK, Yamasaki L and Pagano M . (2003). Dev. Cell, 4, 799–812.
Hattori K, Hatakeyama S, Shirane M, Matsumoto M and Nakayama K . (1999). J. Biol. Chem., 274, 29641–29647.
Hotary K, Allen E, Punturieri A, Yana I and Weiss SJ . (2000). J. Cell Biol., 149, 1309–1323.
Liu J, Stevens J, Rote CA, Yost HJ, Hu Y, Neufeld KL, White RL and Matsunami N . (2001). Mol. Cell, 7, 927–936.
Lustig B and Behrens J . (2003). J. Cancer Res. Clin. Oncol., 129, 199–221.
Matsuzawa SI and Reed JC . (2001). Mol. Cell, 7, 915–926.
Orford K, Crockett C, Jensen JP, Weissman AM and Byers SW . (1997). J. Biol. Chem., 272, 24735–24738.
Polakis P . (2000). Genes Dev., 14, 1837–1851.
Polakis P . (2002). Curr. Biol., 12, R499–R501.
Provost E, Yamamoto Y, Lizardi I, Stern J, D'Aquila TG, Gaynor RB and Rimm DL . (2003). J. Biol. Chem., 278, 31781–31789.
Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW and Pavletich NP . (2003). Mol. Cell, 11, 1445–1456.
Zou JX, Liu Y, Pasquale EB and Ruoslahti E . (2002). J. Biol. Chem., 277, 1824–1827.
Acknowledgements
We thank Hui Zhang, Yale University, Department of Genetics, for providing the HA-Ub construct, Sheng-Cai Lin for the HA-CKIIalpha construct, WJ Harper, Baylor College of Medicine, for the HA-β-TrCP construct, Joseph Madri, Yale University, Department of Pathology, for the use of his Coulter Counter and Addie Tucker for her help with the collagen I assay. Finally, we thank Michael Wolfgang for discussion of some of the experiments and assistance with the figures. Dr Rimm is supported by a grant from the Patrick and Catherine Weldon Donaghue Foundation for Medical Research and grants from the NIH.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Provost, E., McCabe, A., Stern, J. et al. Functional correlates of mutation of the Asp32 and Gly34 residues of beta-catenin. Oncogene 24, 2667–2676 (2005). https://doi.org/10.1038/sj.onc.1208346
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1208346
Keywords
This article is cited by
-
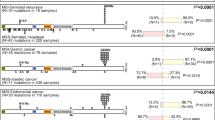
CTNNB1 p.D32A (c.95A > C) somatic mutation in stage I grade 1 endometrioid endometrial carcinoma with lung metastasis: a case report
BMC Medical Genomics (2023)
-
Clinicopathologic and molecular spectrum of testicular sex cord-stromal tumors not amenable to specific histopathologic subclassification
Modern Pathology (2022)
-
Combined activation of MAP kinase pathway and β-catenin signaling cause deep penetrating nevi
Nature Communications (2017)
-
SS18-SSX fusion protein-induced Wnt/β-catenin signaling is a therapeutic target in synovial sarcoma
Oncogene (2014)
-
Analysis of β-catenin alterations in colon tumors: a novel exon 3 mutation
Tumor Biology (2011)