Abstract
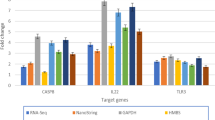
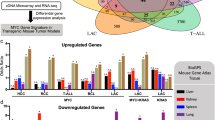
Avian retroviral integration into the c-myb locus is casually associated with the development of lymphomas in the bursa of Farbricius of chickens; these arise with a shorter latency than bursal lymphomas caused by deregulation of c-myc. This study indicates that c-myb mutation in embryonic bursal precursors leads to an oligoclonal population of developing bursal follicles, showing a variable propensity to form a novel lesion, the neoplastic follicle (NF). About half of such bursas rapidly developed lymphomas. Detection of changes in gene expression, during the development of neoplasms, was carried out by cDNA microarray analysis. The transcriptional signature of lymphomas with mutant c-myb was more limited than, and only partially shared with, those of bursal lymphomas caused by Myc or Rel oncogenes. The c-myb-associated lymphomas frequently showed overexpression of c-myc and altered expression of other genes involved in cell cycle control and proliferation-related signal transduction. Oligoclonal, NF-containing bursas lacked detectable c-myc overexpression and demonstrated a pattern of gene expression distinct from that of normal bursa and partially shared with the short-latency lymphomas. This functional genomic analysis uncovered several different pathways of lymphomagenesis by oncogenic transcription factors acting in a B-cell lineage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Abdrakhmanov D, Lodygin D, Geroth P, Arakawa H, Law A, Plachy J, Korn B and Buerstedde J-M . (2000). Genome Res., 10, 2062–2069.
Baba TW and Humphries EH . (1985). Proc. Natl. Acad. Sci. USA, 82, 213–216.
Barth CF and Humphries EH . (1988). Mol. Cell Biol., 8, 5358–5368.
Blume-Jensen P and Humter T . (2001). Nature, 411, 355–365.
Brandvold KA, Ewert DL, Kent SC, Neiman P and Ruddell A . (2001). Oncogene, 20, 3226–3234.
Brandvold KA, Neiman P and Ruddell A . (2000). Oncogene, 19, 2780–2785.
Clurman BE and Hayward WS . (1989). Mol. Cell Biol., 9, 2657–2664.
Clurman BE and Porter P . (1998). Proc. Natl. Acad. Sci. USA 95, 15158–15160.
Cooper MD, Payne LN, Dent PB, Burmester BR and Good RA . (1960). Histogenesis. J. Natt. Cancer Inst., 41, 373–389.
Eckert EA, Beard D and Beard JW . (1951). J. Nat. Cancer Inst., 12, 447–463.
Eisen MB, Spellman PT, Brown PO and Bostein D . (1998). Proc. Natl. Acad. Sci. USA, 95, 14863–14868.
Goitsuka R, Chen CH and Cooper MD . (1997). J. Immunol., 159, 3126–3132.
Gonda TJ, Cory S, Sobieszczuk P, Holtzman D and Adams JM . (1987). J. Virol., 61, 2754–2763.
Gong M, Semus HL, Bird KJ, Stramer BJ and Ruddell A . (1998). J. Vitrol., 72, 5517–5525.
Grandori C, Cowley SM, James LP and Eisenman RN . (2000). Annu. Rev., Cell Dev. Biol., 16, 653–699.
Guarguaglini G, Renzi L, D'Ottavio F, Di Fiore B, Casenghi M, Cundari E and Lavia P . (2000). Cell Growth Diff., 11, 455–465.
Hayward WS, Nell BG and Astrin SM . (1981). Nature 290, 475–480.
Irwin MS and Kaelin Jr WG . (2001). Apoptosis, 6, 17–29.
Jackers P, Minoletti F, Belotti D, Clausse N, Sozzi G, Sobel ME and Castronovo V . (1996). Oncogene, 13, 495–450.
Jiang W, Kanter MR, Dunkel I, Ramsay RG, Beemon KL and Hayward W.S . (1997). J. Vitrol., 71, 6526–6533.
Kanter MR, Smith RE and Hayward WS . (1988). J. Vitrol., 62, 1423–1432.
Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M and Peters JM . (2000). Mol. Biol. Cell, 11, 1555–1569.
Kuersten S, Mutsuhito O and Mattaj I. W . (2001). Trends Cell Biol., 11, 497–503.
Leprince D, Gegonne A, Coll J, Taisne C, Schneederger A, Lagrou C and Stehelin D . (1983). Nature, 306, 395–397.
Marshall AJ, Niiro H, Lerner CG, Yun TJ, Thomas S, Disteche CM and Clark EA . (2000). J, Exp. Med., 191, 1319–1332.
Muzi-Falconi M and Kelly TJ . (1995). Proc. Natl. Acad. Sci. USA, 92, 12475–12479.
Nason-Burchenal K and Wolff L . (1993). Proc. Natl. Acad. Sci, USA, 90, 1619–1623.
Neel BG, Hayward WS, Robinson HL, Fang J and Astrin SM . (1981). Cell, 23, 323–334
Neiman P, Payne LN and Weiss RA . (1980). J, Virol., 34, 178–186.
Neiman PE . (1994). Adv. Immunol., 56, 467–484.
Neiman PE, Ruddell A, Jasoni C, Loring G, Thomas SJ, Brandvold KA, Lee R, Burnside J and Delrow J . (2001). Proc. Natl. Acad. Sci. USA, 98, 6378–6383.
Neiman PE, Thomas SJ and Loring G . (1991). Proc. Natl. Acad. Sci. USA, 88, 5857–5861.
Neiman P, Wolf C, Enrietto PJ and Cooper GM . (1985). Proc. Natl. Acad. Sci. USA, 82, 222–236.
Ness SA . (1996). Biochim, Biophys. Acta 1288, F123–F139.
Ouspenski II . (1998). Exp. Cell Res., 224, 171–183.
Payne GS, Courtneidge SA, Crittenden LB, Fadly AM, Bishop JM and Varmus HE . (1981). Cell, 23, 311–322.
Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters JM and Helin K . (2000). Genes Dev., 14, 2330–2343.
Pfleger CM, Lee E and Kirschner MW . (2001). Genes Dev., 15, 2396–2407.
Pizer ES, Baba TW and Humphries EH . (1992). J. Vitrol., 66, 512–523.
Pizer E and Humphries EH . (1989). J. Virol., 63, 1630–1640.
Prichard CC, Li H, Delrow J. and Nelson PS . (2001). Proc. Natl. Acad. Sci. USA, 98, 13266–13271.
Sauer K, Lior J, Singh SB, Yablonski D, Weiss A and Perlmutter RM . (2001). J. Biol. Chem., 276, 45207–45216.
Schmidt M, Nazarov V, Stevens L, Watson R and Wolff L . (2000). Mol. Cell. Biol., 20, 1970–1981.
Shen-Ong GL, Morse III HC, Potter M and Mushinski JF . (1986). [Published erratum appears in Mol. Cell Biol. 1986; 6: 2756]. Mol. Cell Biol., 6, 380–392.
Shen-Ong GL and Wolff L . (1987). J. Virol., 61, 3721–3725.
Smith MR, Smith RE, Dunkel I, Hou V, Beemon KL and Hayward WS . (1997). J. Virol., 71, 6534–6540.
Sudo T, Ota Y, Kotani S, Nakao M, Takami Y, Takeda S and Saya H . (2001). EMBO J., 20, 6499–6508.
Thompson CB, Humphries EH, Carlson LM, Chen C-LH and Neiman PE . (1987). Cell, 51, 371–381.
Tirunagaru VG, Sofer L, Cui J and Burnside J . (2000). Genomics, 66, 144–151.
Wolff L, Mushinski JF, Shen-Ong GL and Morse III HC . (1988). J. Immunol., 141, 681–689.
Acknowledgements
The authors thank Alanna Ruddell for helpful comments and Joan Burnside, Jean-Marie Buerstedde and the Resource Center of the German Genome Project for their cooperation in obtaining cDNA clones for the microarray and subsequent analyses. This work was supported by NIH grants RO1 CA20068 to PEN and RO1 CA48746 to KLB.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Neiman, P., Grbiç, J., Polony, T. et al. Functional genomic analysis reveals distinct neoplastic phenotypes associated with c-myb mutation in the bursa of Fabricius. Oncogene 22, 1073–1086 (2003). https://doi.org/10.1038/sj.onc.1206070
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1206070
Keywords
This article is cited by
-
Kruppel-Like Factor KLF4 Facilitates Cutaneous Wound Healing by Promoting Fibrocyte Generation from Myeloid-Derived Suppressor Cells
Journal of Investigative Dermatology (2015)
-
Mutation of an IKK phosphorylation site within the transactivation domain of REL in two patients with B-cell lymphoma enhances REL's in vitro transforming activity
Oncogene (2007)
-
Myb proteins inhibit fibroblast transformation by v-Rel
Molecular Cancer (2006)
-
Genomic instability during Myc-induced lymphomagenesis in the bursa of Fabricius
Oncogene (2006)
-
Development of a cDNA array for chicken gene expression analysis
BMC Genomics (2005)