Abstract
We describe the use of pooled, region-specific hybridisation probes to screen high-density replica filters of a human genome YAC library. The probes were derived by microdissection of an approximately 30-Mbp region subtending the translocation breakpoint on a der(1)(1;11)(q42.1;q14.3) chromosome. Of 70 microdissection clones used in pools of 4–10, 47 identified a total of 77 YAC recombinants, representing over 50% of the microdissected region. This strategy can easily be adapted to other poorly mapped subchromosomal regions of the human or other mammalian genomes and will provide a solid framework for detailed contig map constructions.
Similar content being viewed by others
Introduction
Large-scale genome mapping projects have benefited enormously from the ability to clone large fragments of genomic DNA as artificial chromosomes (YACs) in the budding yeast Saccharomyces cerevisiae [1]. This technology allows coverage of large chromosomal regions or even entire chromosomes with a relatively limited number of overlapping clones [2, 3].
Contig mapping generally relies on a sufficient density of markers to anchor clones on the chromosomal map. However, certain regions of the genome are poorly mapped because of a relative dearth of markers, therefore constraining chromosome walking and contig mapping exercises. It is then desirable to produce new markers and to screen largeinsert genomic libraries with many markers at once to accelerate contig mapping.
St. Clair et al. [4] described a Scottish family with a balanced t(1;11)(q42.1;q14.3) translocation which segregates with schizophrenia. As part of a strategy towards cloning the translocation breakpoint, we produced a large number of new markers by microdissecting and microcloning regions 1q41-q42.3 and 11q14.3-q23 from the der(1) translocation chromosome of one of the patients [5]. FISH painting performed with the whole library of microdissection clones (MDs) showed that the signal mapped equally to chromosomal regions 1q41-q42.3 and 11q14.3-q23[5].
We screened high-density replica filters [6] of a whole human genome library [7] by hybridisation with pools of 4–10 repeat-free MDs, leading to the isolation of 162 YACs. We describe the analysis of this region-specific YAC minilibrary, and discuss the efficacy of the technique as well as its practicability as a general approach to initiating contig maps of large genomic regions.
Materials and Methods
Primary Screening
High-density replica filters of the ICI library [7] were produced at the Human Gene Mapping Project Resource Centre in Harrow (UK) as described elsewhere [6]. Inserts from the MDs were amplified by PCR using vector-specific primers [8, 9]. Insert DNA was carefully quantified by agarose gel electrophoresis followed by ethidium bromide staining. Pools of 4–10 MD inserts were made by mixing equal amounts of DNA from individual inserts to normalise the hybridisation of each MD as far as possible. 20 ng of pool DNA were radiolabelled [10] and hybridised to the ICI high-density replica filters as described elsewhere [6], except from pool 7 which required repeat suppression with human Cotl DNA (Gibco BRL; performed following the supplier’s instructions). Filters were then washed as described in Bentley et al. [6] and exposed for autoradiography to Kodak XAR films between two intensifying screens at −80°C for 1–5 days.
Secondary Screening of the Minilibrary
The YAC clones identified in the primary screening were ordered in two 96-well microtitre plates which were replicated onto Hybond N (Amersham) using a 96-prong device [11], grown on AHC agar for 48 h, and processed [6], using Glucanex (Novo Nordisk Ferment) as a yeast lytic enzyme [12]. Secondary screening was performed with individual, radiolabelled MDs [10].
Southern blot analysis of the YACs was performed by digesting 500 ng of total yeast DNA with EcoRI (Eurogentec), followed by electrophoresis through a 0.9% agarose gel and transfer onto Hybond N. Probe labelling, hybridisation and washes were as described for the primary screening.
Results
Primary Screening of a Human YAC Library
Inserts of 600 phages obtained by microdissecting the chromosomal segment encompassing the t(1; 11)(q42.1;q14.3) translocation breakpoint were screened for the presence of repetitive sequences by Southern blot analysis with a total human DNA probe. 70 repeatfree MDs were identified. The insert size ranged between 500 bp and 4 kbp, with an average size of 1.5 kbp. Ten complex probes were prepared by mixing between 4 and 10 MDs (table 1), radiolabelled [10] and hybridised to high-density replica filters of a YAC library [7].
As screening with complex probes was expected to result in non-uniform signals from between MDs within a pool, some clones giving signals just above background were picked, although strongly hybridising clones were selected preferentially.
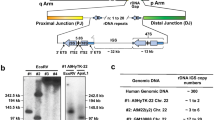
Typical results are shown in figure 1. Panel A shows one filter screened with pool 3 (table 1). All four strongly hybridising clones (numbered 1–4) proved to be real positives by Southern blot analysis (see ‘Secondary Screening’). Panel C shows a filter from screening with pool 8. Clones labelled 1–4 were picked, but only clone 1 was found positive at later stages of analysis (data not shown).
Results from screening with the 10 pools are indicated in table 1. A minilibrary of 162 YAC clones was collected.
Secondary Screening of the Minilibrary
Secondary screening of the minilibrary was performed by hybridisation with individual MD clones. A YAC was considered genuine when it gave, on a Southern blot, an EcoRI hybridisation signal of the same size as in total human genomic DNA and in the EcoRI-cloned MD insert. Southern blot analysis of some of the clones identified in the primary screening with pool 3 is displayed in figure 1b, and shows that clones 1CG5 and 25EH3, and clone 1CF2 are positive for MD21 and MD27, respectively. In total, 77 YACs out of 162 (47.5%) were found to contain one or more MDs and were therefore considered to be real positives.
47 MDs out of 70 (67%) used in the primary screening identified at least one positive YAC in the minilibrary, and will be referred to hereafter as ‘positive MDs’. This proportion is as high as could be expected, considering the fact that the probes had not been characterised prior to undertaking the screening, except that they were known to be repeat free. Positive MDs detected between one and six YACs in the minilibrary.
The minilibrary was further evaluated by screening with several markers known to map to the microdissected chromosomal segments. Positive YACs were found for markers D1S103 [13] and D1S251 [14] on chromosome 1, and for the ionotropic glutamate receptor gene GLUR4 and marker D1S351 on chromosome 11 [15]. However, no YAC was found at loci STMY and D11S388 although markers mapping both proximal and distal to these markers are known to exist in the MD library [18].
Effect of Pool Size, Insert Size and Presence of Moderately Iterated Repeats on the Screening
The rate of genuine positive YACs as well as the proportion of positive MDs vary considerably from one pool to another (table 1). The effect of three parameters was examined.
Number of MDs per Pool
Although a slight trend towards lower yields was observed with the larger pools, no major difference between the pools was observed in the proportions of either positive MDs or positive YACs.
Insert Size
Whereas the insert size did not seem to have a major effect on the percentage of positive MDs, an obvious trend towards higher rates of false-positive YACs was observed with the larger inserts. 60–70% of genuine positive YACs were obtained with insert sizes up to 1.6 kbp (pools 2, 3, 4 and 9; the latter contains inserts with sizes mostly between 0.5 and 1.3 kbp). The figures dropped consistently with inserts larger than 1.6 kbp, down to 35 and 20% for pool 7 and 8, respectively. This showed that insert size had a major effect on the screening.
Presence of Moderately Iterated Repeats in the Inserts
As indicated in table 1, primary screening with pools 5,7, 8 and 10 produced a significant level of hybridisation background and led to high rates of false-positive YACs. Secondary screening with individual MDs showed that each of these pools contained an MD with a moderately iterated repeat in its insert. The vast majority (60–100%) of falsepositive YACs obtained with these pools hybridised in an aspecific manner to these particular MDs, indicating that the repeat-containing MDs were responsible for the high rate of false positive YACs in the corresponding pools. Indeed, pool 1 initially contained an MD with a repetitive element, and no genuine positive YAC was identified. When primary screening was repeated without this MD, 41% of YACs identified in the primary screening were genuine.
Discussion
We have used 70 MDs derived from regions 1q41-q42.3 and 11q14.3-q23 pooled in sets of 4–10 to screen high-density replica filters of a human genome YAC library. We isolated 162 YACs, 77 of which (47.5%) were found to be positive for one or more MDs in the secondary screening. Of the 70 MDs, 47 (67%) identified YACs.
The size of the inserts played a major role on the outcome of the screening. The percent-age of genuine positive YACs was consistently above 60% as long as the individual probe sizes remained below 1.6 kbp, but dropped as soon as they exceeded 1.6 kbp. The presence of moderately iterated sequences in four inserts accounted for the vast majority of falsepositive YACs observed in our experiment.
The MDs were assigned to either chromosome 1 or chromosome 11 using a panel of somatic hybrid cell lines [8, 16]. 23 were assigned to chromosome 1, and 33 to chromosome 11. The remaining 14 MDs could not be assigned to either chromosome unambiguously.
On the basis of the average insert size of the YACs in the library (350 kbp), the 36 YACs containing MDs assigned to chromosome 1 and the 30 YACs containing MDs assigned to chromosome 11 cover a total of 12,600 and 10,500 kbp, respectively. The 11 YACs corresponding to the MDs that could not be assigned to either chromosome covered an estimated extra 3,850 kbp of DNA.
The 77 YACs identified in this work are anchored at 47 MDs. The microdissected segment is estimated at about 30 Mbp, giving an average density of one positive MD per 650 kbp. Given that the mean size of the YACs is 350 kbp, it is likely that about half of the microdissected region is covered with YACs. Although the repartition of MDs follows a Gaussian distribution centered around the t(1; 11) translocation breakpoint [5,8], one can estimate that our minilibrary of YACs constitutes the backbone for constructing a contig map of the whole microdissected segment.
Confirmation and closure of the map will require further screening with additional, selected MDs or STS markers and will benefit from the emerging whole genome genetic and physical map [17]. Assignment of the chromosome 11q MDs to nine subregions defined by a panel of somatic hybrid cell lines [8, 16] allowed the direct physical localisation of the YACs which contain these MDs. This will facilitate the elaboration of an integrated map of these chromosomal intervals.
We propose that a strategy in which MDs are used to screen large-insert genomic libraries and which are subregionally mapped with a panel of reduced somatic hybrid cell lines will be a powerful procedure to initiate the construction of a contig map in any region of the genome.
References
Burke DT, Carle GF, Olson MV: Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science 1987;236:806–812
Chumakov I, Rigault P, Guillou S, Ougen P, Billaut A, Guasconi G, Gervy P, LeGall I, Soularue P, Grinas L, Bougueleret L, Bellanné-Chantelot C, Lacroix B, Barillot E, Gesnouin P, Pook S, Vaysseix G, Frelat G, Schmitz A, Sambucy JL, Bosch A, Estivill X, Weissenbach J, Vignal A, Riethman H, Cox D, Patterson D, Gardiner K, Hattorf M, Sakaki Y, Ichikawa H, Ohki M, Le Paslier D, Heilig R, Antonarakis S, Cohen D: Continuum of overlapping clones spanning the entire human chromosome 21q. Nature 1992;359:380–387
Foote S, Vollrath D, Hilton A, Page DC: The human Y chromosome: Overlapping DNA clones spanning the euchromatic region. Science 1992;258:60–66
St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, Gosden C, Evans HJ: Association within a family of a balanced autosomal translocation with major mental illness. Lancet 1990;336:13–16
Muir WJ, Gosden CM, Brookes AJ, Fantes J, Evans KL, Maguire SM, Stevenson B, Boyle S, Blackwood DHR, St Clair DM, Porteous DJ, Weith A: Direct microdissection and microcloning of a translocation breakpoint region t(1;11) (q42.2; q21) associated with schizophrenia. Cytogenet Cell Genet 1995;70:35–40
Bentley DR, Todd C, Collins J, Holland J, Dunham I, Hassock S, Bankier A, Giannelli F: The development and application of automated gridding for efficient screening of yeast and bacterial ordered libraries. Genomics 1992;12:534–541
Anand R, Riley JH, Smith JC, Markham AF: A 3.5 genome equivalent multi access YAC library: Construction, characterisation, screening and storage. Nucleic Acids Res 1990;18:1951–1955
Evans KL, Brown J, Shibasaki Y, Devon RS, He L, Arveiler B, Christie S, Maule JC, Baillie D, Slorach EM, Anderson SM, Gosden JR, Petit J, Weith A, Gosden CM, Blackwood DHR, St Clair DM, Muir WJ, Brookes AJ, Porteous DJ: A contiguous clone map over 3Mbp on the long arm of chromosome 11 across a balanced translocation associated with schizophrenia. Genomics 1995;28:420–428
Gussow D, Clackson T: Direct clone characterization from plaques and colonies by the polymerase chain reaction. Nucleic Acids Res 1989; 17:4000.
Feinberg AP, Vogelstein B: A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 1983;132:6–13
Arveiler B: Construction of chromosome-specific libraries of yeast artificial chromosome recombinants from somatic hybrid cell lines; in Gosden JR (ed): Methods in Molecular Biology. Totowa, Humana Press 1994, vol 29: Chromosome Analysis Protocols.
Petit J, Boisseau P, Arveiler B: Glucanex: A cost effective yeast lytic enzyme. Trends Genet 1994;10:4–5
Engelstein M, Hudson TJ, Lane JM, Lee MK, Leverone B, Landes GM, Peltonen L, Weber JL, Dracopoli NC: A PCR-based linkage map of human chromosome 1. Genomics 1993;15:251–258
Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J: The 1993–94 Généthon human genetic linkage map. Nat Genet 1994;7:246–339
James MR, Richard CW III, Schott JJ, Yousry C, Clark K, Bell J, Terwilliger JD, Hazan J, Dubay C, Vignal A, Agrapart M, Iami T, Nakamura Y, Polymeropoulos M, Weisenbach J, Cox DR, Lathrop GM: A radiation hybrid map of 506 STS markers spanning human chromosome 11. Nat Genet 1994;8:70–76
Evans KL, van Heyningen V, Porteous DJ: Placement and refined mapping of established and new markers on human chromosome 11q using a small panel of somatic cell hybrids. Eur J Hum Genet 1995;3:42–48
Cohen D, Chumakov I, Weissenbach J: A first-generation physical map of the human genome. Nature 1993;366:698–701
Acknowledgements
We are grateful to Sarah Smith and Ramnath Elaswarapu (Human Genome Mapping Project Resource Centre, Cambridge, UK) for help in making the highdensity replica filters, and for efficient YAC supply. This work was supported by grants from the Fondation pour la Recherche Médicale, Groupement de Recherche et d’Etude sur les Génomes, and the Conseil Régional d’Aquitaine to B.A., from the Medical Research Council Human Genome Mapping Project to D.P. and from the Wellcome Trust Foundation to D.StC.
Author information
Authors and Affiliations
Additional information
The first two authors made an equal contribution to this work.
Rights and permissions
About this article
Cite this article
Petit, J., Boisseau, P., Evans, K. et al. Seeding of YACs over Regions 1q41-q42.3 and 11q14.3-q23 with Microdissection Clones. Eur J Hum Genet 3, 351–356 (1995). https://doi.org/10.1159/000472324
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472324