Abstract
In vitro studies in bacterial, yeast and eukaryotic systems have demonstrated the existence of deletion and insertion ‘hotspots’ involving repetitive sequences. Slipped-strand mispairing (SSM) has been suggested to be the mechanism involved. Progress in human molecular genetics has allowed the identification of many mutations causing diseases. Analysis of sequences involved in these mutations provides an opportunity to investigate the contribution of short tandem repeats to the naturally occurring mutations in coding regions of human genes. We have analyzed the sequences surrounding 625 disease-causing mutations in the coding regions of three genes: the cystic fibrosis transmembrane conductance regulator, β globin and factor IX. Altogether, 134 (21%) insertion and deletion mutations of 4 base pairs or less were identified. In 47% of these mutations, the deletions and insertions occurred within a unit repeated tandemly 2- to 7-fold. These were classified as SSM mutations. The proportion of SSM mutations was significantly higher than expected by chance. The estimated net proportion of deletion and insertion mutations attributed to SSM was 27%. These results indicate that very short repetitive sequences contribute significantly to the generation of deletion and insertion mutations in human genes, and to the evolution of diversity of their coding regions.
Similar content being viewed by others
Introduction
Genetic information can be altered by base substitutions or by addition or deletion of nucleotides. These changes can be either beneficial, neutral or detrimental to the organism. In order to understand the mechanisms generating mutations, it is essential to investigate the nature of the DNA sequence alterations. Extensive studies in bacterial genetic systems have demonstrated the existence of deletion and insertion ‘hotspots’, involving repetitive sequences [1, 2]. In vitro frameshift fidelity assays using eukaryotic DNA polymerases have suggested that template-primer misalignment during DNA synthesis or recombination is probably the mechanism generating short deletion and insertion mutations [3], According to this proposal, misaligned intermediates are formed as a result of slippage of DNA strands, in regions containing repeated nucleotides. Therefore, the mechanism was termed slipped-strand mispairing (SSM) [4].
The eukaryotic genome contains many regions in which small motifs consisting of a single base or small number of bases are repeated in tandem multiple times (often over 20 repeated units in a run). These relatively high-copy-number simple tandem repeats comprise the highly polymorphic microsatellite sequences found in many noncoding regions of mammalian genomes. These polymorphisms derive from differences in the copy number of the tandemly repeated simple motifs, and are usually stably inherited. Therefore, these polymorphic sites are useful for genomic mapping and for fingerprinting studies. Analysis of several restriction fragment length polymorphism (RFLP) sites flanking polymorphic microsatellite sites has suggested that SSM, rather than unequal exchange between homologous chromosomes, is most probably the mechanism involved in generating copy number variation [5,6].
Studies in Saccharomyces cerevisiae have shown that (GT)n tracts are highly unstable, with length alterations at a minimal rate of 10−4 events per division [7]. Most of the changes involve additions or deletions of one or two repeated units. In addition, Strand et al. [8] have shown that the instability of poly (GT) sequences in yeast can be greatly increased by mutations in DNA mismatch repair genes. These results support the assumption that tract instability is associated with DNA polymerases slipping during replication. Another study has recently shown that DNA plasticity in mitochondrial DNA is a result of the instability of a 10-bp tandemly repeated sequence [9].
Several human diseases (fragile X, myotonic dystrophy, Huntington’s chorea, spinal and bulbar muscular atrophy and spinocerebellar ataxia type 1) have been found to be caused by somatic expansion of highly repeated tandem repetitive sequences [10–15]. In these diseases, a dramatic increase in the length of the repeated run occurs.
Recent progress in human molecular genetics has led to the identification of disease-causing mutations in several human genes. Analysis of the sequences involved in these mutations has provided an opportunity to investigate the mechanisms involved. In a study of 80 short deletion and insertion mutations, direct repeats of between 2 and 8 bp were found in the immediate vicinity of the majority of the analyzed mutations [16, 17]. These direct repeats either included or partially overlapped the deleted or inserted bases. A modified SSM model was proposed to explain these results. The existence of direct repeats in the vicinity of mutation sites is not surprising, since an extensive computer research of DNA sequences has shown significantly high levels of nontandem direct repeats (cryptic simplicity) in many coding or non-coding DNA sequences [18].
In this study, a more conservative analysis was performed in order to estimate the net influence of SSM in short (1–4 bp) insertion and deletion mutations. This was obstained by classifying a mutation as SSM only in those cases where the mutation could be simply explained by the SSM model as detailed in the Material and Methods section. The results of this analysis are expected to enhance our understanding of the contribution of SSM to the mutability of human genes. We have studied three genes in which a large number of disease-causing mutations have been identified: (1) the cystic fibrosis (CF) transmembrane conductance regulator (CFTR) gene, in which 400 mutations have been identified [CF Genetic Analysis Consortium, pers. commun.]; (2) the β globin gene, in which over 100 mutations cause β thalassemia [19], and (3) the factor IX gene, in which over 300 mutations cause hemophilia B [20].
Materials and Methods
Sequences flanking mutations in the coding region of the CFTR, β-globin and factor IX genes were analyzed. The information was obtained from previously published reports [19, 20; CF Genetic Analysis Consortium, pers. commun.]. The analyzed mutations were located within published normal coding sequences as reported in GenBank.
Classification of Mutations
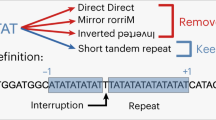
Mutations that were neither insertions nor deletions, and mutations in which the insertion or deletion was larger than 4 bp were classified as ‘other’ mutations. Classification as an SSM mutation was restricted to mutations of up to 4 bp since most length variation was found in repeat units of 1–4 bp [21]. Insertion or deletion mutations of 4 bp or less were classified as deriving from SSM if they met the following conditions: (1) deletion of n (1–4) bp was considered SSM, if and only if the adjacent n bases, from either side, were identical to the deleted bases; (2) insertion of n (1–4) bp was considered SSM if and only if the insertion was adjacent to a tandem repeat of at least 2 repeats with the same sequence as the inserted bases, and (3) when the deletion or insertion was itself a repetitive unit (e.g. TT, ACAC), only the repeat unit was compared with the adjacent bases to classify the nature of the mutation according to (1) and (2) (e.g. deletion of AA in a run of AAA would be considered SSM).
All other mutations were classified as non-SSM. At each SSM mutation, the length of the repetitive run in the normal sequence was counted. For example: (1) deletion of the underlined nucleotides in the normal sequence ACACACAC was counted as a 2-bp deletion SSM mutation in a run of four repeats and (2) insertion of the double underlined nucleotide in the sequence AAA was counted as a 1-bp insertion SSM mutation in a run of two repeats.
It is important to note that deletion and insertion mutations might be classified as SSM if they happened to occur adjacent to a repetitive sequence, even if SSM is not actually the mechanism responsible for the mutation. Under the assumption that SSM is not involved in generating deletion and insertion mutations, the proportion of deletion and insertion mutations expected to be classified as SSM by chance was calculated as follows: (1) Deletion mutations. The proportion of 1-bp deletions was calculated by counting all the base pairs that if deleted would have been considered to result from SSM, divided by the total number of base pairs in the coding sequence analyzed. The proportions of all possible 2-, 3- and 4-bp deletions that would be classified as SSM were similarly calculated. (2) Insertion mutations. The proportion of 1-bp insertions was calculated by counting all the locations in which a 1-bp insertion would have been considered to result from SSM, and dividing by the total number of possible locations for insertions (the total number of base pairs + 1). The inserted nucleotides were taken to be A, C, G or T, each with equal probability. The proportion of 2-, 3- and 4-bp insertions was calculated in a similar manner by considering all possible 2, 3 and 4 bp at each point along the sequence. The net proportion of mutations that can be attributed to the mechanism of SSM was calculated as described in the Appendix.
Results
Several examples of the classification of mutations are presented in table 1. Table 2 presents the total number of mutations analyzed for each gene, the total number of insertion and deletion mutations and the proportion of the insertion and deletion mutation that can be classified as SSM mutations. The proportion of the total number of mutations that were insertions or deletions varied from 0.13 for factor IX to 0.45 for β globin. The proportion of total insertion and deletion mutations classified as SSM was 0.47.
Table 3 presents results of the analysis of insertion and deletion mutations according to the number of deleted or inserted base pairs. Deletion and insertion mutations were analyzed separately. Results are presented for the three genes together, since similar proportions were found independently in each gene (data not shown). The number of deletion mutations (100) was 3-fold greater than the number of insertion mutations (34). In cases in which SSM mutations were present, the proportions of SSM mutations were tested for statistical difference from the proportions expected by random chance using an exact binomial test. With one exception, there were significantly more SSM mutations than would be expected by chance. The exception was the 1-bp deletions, for which the proportion was not significantly greater than that expected by chance (0.57 versus 0.48). The net proportion of mutations attributed to SSM was greater in insertions (0.36) than in deletions (0.24), with an overall proportion of 0.27.
Table 4 presents the distribution of SSM insertion and deletion mutations, according to the number of repeats in the run. Mutations of 1 bp and mutations of > 1 bp were analyzed separately. It can be seen that the most frequent deletion mutation event is a 1-bp deletion in a run of two units. These mutations, however, are attributed mostly to a mechanism other than SSM, and classified as SSM only by chance. Insertion mutations are more common in longer runs.
Discussion
The study analyzed 625 different disease-causing mutations in the coding regions of three human genes: CFTR, β globin and factor IX. A strict analysis was performed in order to estimate the net influence of SSM in short (1–4 bp) insertion and deletion mutations. This was obtained by classifying a mutation as SSM only in those cases in which the mutation could be simply explained by the SSM model. It is important to note that the repertoire of mutations found in patients showing disease symptoms does not reflect the entire spectrum of mutational events that might have occurred in these coding sequences in the course of evolution. Mutations without clinical effect or mutations that have not ‘survived’ through evolution are missing. The search uncovered 134 deletion or insertion mutations of 4 bp or less (table 2). These were 21% of all mutations. Our analysis revealed that a net proportion of 27% of all deletion or insertion mutations of 4 bp or less can be attributed to SSM events. In these cases, it is reasonable to assume that SSM, rather than unequal sister chromatid exchange, is the mechanism involved, since the analyzed mutations are within very short runs (2–7 repeats) which probably would not facilitate unequal pairing of DNA strands.
As seen in table 3, the proportion of deletion and insertion mutations that can be explained by SSM is significantly higher than expected by chance, except for the 1-bp deletions. These results indicate that SSM is a significant mechanism causing deletion and insertion mutations in human genes. The appearance of a simple run of 2 bp (AA, CC, GG and TT) is the most common repeat in any sequence. Therefore, it is not surprising that, as seen in table 4, the majority of the 1-bp deletion mutations are in runs of two repeats. This supports the suggestion that some of these mutations are caused by other mechanisms and are classified as SSM by chance only.
It is well recognized that the highly polymorphic microsatellite regions comprised of long runs (>20) of short tandem repeats are unstable, leading to changes in the number of the repeated unit. In addition, several human diseases have been found to be caused by the instability of long runs of 3 bp tandemly repeated. Our results suggest that very short runs (<8) of tandem repeats can also lead to misalignment and SSM. Since the frequency of short tandem repeats is higher than the frequency of long tandem repeats, the results presented here indicate that the instability of these short repeats contributes to DNA mutability in human genes.
Nontandem repeats, interrupted by a few base pairs, may also promote deletion and insertion mutations by SSM. This has been convincingly demonstrated in Escherichia coli [25, 26]. In our study, such mutations were not classified as SSM mutations. Nevertheless, several examples have been observed, such as a 4-bp deletion (double underlined) in the CF gene: CTACCAAGTCAACCAAACCATACAA-3667del4 [Estivill, pers. commun.]. This sequence comprises three repetitive units (underlined) of 4 bp, interrupted by a few base pairs. Thus, the proportion of deletion and insertion mutations in which short repeats are involved may well be even higher than found in this study. Furthermore, it has been suggested that the SSM mechanism is also involved in generating substitution mutations [27]. Thus, the SSM mechanism is probably a major mechanism for the generation of mutations in general.
References
Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M: Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol 1966;31:77–84
Halhday JA, Glickman BW: Mechanisms of spontaneous mutation in DNA repair-proficient Escherichia coli. Mutat Res 1991;250:55–71.
Runkel TA: Frameshift mutagenesis by eukaryotic DNA polymerases in vitro. J Biol Chem 1986;261:13581–13587
Levinson G, Gutman G: Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol Biol Evol 1987;4:203–221
Morral N, Nunes V, Casals T, Estivill X: CA/GT microsatellite alleles within the cystic fibrosis transmembrane conductance regulator (CFTR) gene are not generated by unequal crossing over. Genomics 1991;10:692–698
Wolf R, Plaetke R, Jeffreys AJ, White R: Unequal crossing over between homologous chromosomes is not the major mechanism involved in the generation of new alleles at VNTR loci. Genomics 1989;5:382–384
Henderson ST, Petes TD: Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol 1992;12:2749–2757.
Strand S, Prolla TA, Liskay RM, Petes TD: Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 1993;365:274–276
Madsen CS, Ghivizzani SC. Hauswirth WW: In vivo and in vitro evidence for slipped mispairing in mammalian mitochondria. Proc Natl Acad Sci USA 1993;90:7671–7675.
Fu YH, Kuhl DPA, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJMH, Holden JJA, Fenwick RG Jr, Warren ST, Oostra BA, Nelson DL, Caskey CT: Variation of the CGG repeat at the fragile X site results in genetic instability: Resolution of the Sherman paradox. Cell 1981;67:1047–1058
Kremer EJ, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren ST, Schlessinger D, Sutherland GR, Richards RI: Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science 1991;252:21711–21714
Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, Sohn R, Zemelman B, Snell RG, Rundle SA, Crow S, Davies J, Shelbourne P, Buxton J, Jones C, Juvonen V, Johnson K, Harper PS, Shaw DJ, Housman DE: Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 1992;68:799–808
The Huntington’s Disease Collaborative Research Group: A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993;72:971–983
La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH: Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 1991;352:77–79.
Orr HT, Chung MY, Banfi S, Kwiatkowski TJ Jr, Servadio A, Beaudet AL, McCall AE, Duvick LA, Ranum LPW, Zoghbi HY: Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nature Genetics 1993;4:221–226
Krawczak M, Cooper DN: Gene deletions causing human genetic disease: Mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum Genet 1991;86:425–441
Cooper DN, Krawczak M: Mechanisms of insertional mutagenesis in human genes causing genetic disease. Hum Genet 1991;87:409–415
Tautz D, Trick M, Dover GA: Cryptic simplicity in DNA is a major source of genetic variation. Nature 1986;322:652–656
Kazazian HH Jr: The thalassemia syndromes: Molecular basis and prenatal diagnosis in 1990. Semin Hermatol 1990;27:209–228
Giannelli F, Green PM, High KA, Sommer S, Poon MC, Ludwig M, Schwaab R, Lozier JN, Reitsma PH, Goossens M, Yoshioka A, Brownlee GG: Haemophilia B: Database of point mutations and short additions and deletions. Nucleic Acids Res 1993;21:3075–3087
Vogt P: Potential genetic function of tandem repeated DNA sequence blocks in the human genome are based on a highly conserved ‘chromatin folding code’. Hum Genet 1990;84:301–336
Green PM, Bentley DR, Mibashan RS, Nilsson IM, Giannelli F: Molecular pathology of haemophilia B. EMBOJ 1989;8:1067–1072
Zielenski J, Bozon D, Kerem B, Markiewicz D, Rommens JM, Tsui LC: Identification of mutations in exons 1 through 8 of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics 1991;10:229–235
Iannuzzi MC, Stern RC, Collins FS, Tom Hon C, Hidaka N, Strong T, Becker L, Drumm M, White MB, Gerrard B, Dean M: Two frameshift mutations in the cystic fibrosis gene. Am J Hum Genet 1991;48:227–231
Albertini AM, Hofer M, Calos MP, Miller JH: On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell 1982;29:319–328
Balbinder E, MacVean C, Williams RE: Overlapping direct repeats stimulate deletions in specially designed derivatives of plasmid pRB325 in Escherichia coli. Mutat Res 1989;214:233–252.
Fieldhouse D, Golding B: A source of small repeats in genomic DNA. Genetics 1991;129:563–572
Acknowledgements
Profound thanks to Prof. Morris Soller for his valuable comments. This work was supported by grants from the Thyssen Foundation (Germany) and the National Academy for Sciences (Israel).
Author information
Authors and Affiliations
Appendix
Appendix
As explained in the text, a mutation of another nature that occurred by chance within a repetitive sequence, would be classified as SSM. In order to estimate the net proportion of mutations that can be explained by a SSM event, Pts, the following procedure was carried out. The analysis is done separately for each type of mutation as defined in table 3 (i.e., deletion or insertion and the number of base pairs involved). The following definitions are used:
-
MALL = The total number of mutations.
-
McS = The number of mutations that were classified as SSM.
-
MtS = The estimated number of mutations that were caused by a true SSM event.
-
McO = The number of mutations that were classified as other than SSM mutations.
-
MtO = The estimated number of mutations that were caused by a non-SSM event.
-
PcS = The proportion of mutations that were classified as SSM.
-
PrS = The proportion of mutations that would be classified as SSM in the given sequence as a result of a random process generating the mutations.
-
PtS = The estimated proportion of mutations that were caused by a true event of SSM.
-
McS and McO can then be expressed as: McS = MtS + PrSMtO, and McO = MtO − PrSMtO. These two equations have two unknown parameters, MtS and MtO. Therefore, a solution for MtS can be obtained: MtS = McS − [PrSMcO/(1 − PrS)]. Pts can then be obtained: PtS = MtS/MALL.
Rights and permissions
About this article
Cite this article
Darvasi, A., Kerem, B. Deletion and Insertion Mutations in Short Tandem Repeats in the Coding Regions of Human Genes. Eur J Hum Genet 3, 14–20 (1995). https://doi.org/10.1159/000472269
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472269
Key Words
This article is cited by
-
In human genome, generation of a nuclear export signal through duplication appears unique to nucleophosmin (NPM1) mutations and is restricted to AML
Leukemia (2008)
-
New splicing mutations in propionic acidemia
Journal of Human Genetics (2006)