Abstract
We have used eight PCR-based DNA polymorphisms to determine the parental origin and mechanisms of formation in 9 patients with de novo nonmosaic tetrasomy 18p. The 9 patients, 4 girls and 5 boys, had clinical features characteristic of i(18p) syndrome. The supernumerary marker chromosome was identified by fluorescence in situ hybridization (FISH) analysis using centromeric probes and a flow-sorted 18p-specific library. The isochromosome was of maternal origin in all 9 cases. The formation of tetrasomy 18p cannot be explained by a single model. In 6 cases, meiosis II nondisjunction, followed by subsequent postzygotic misdivsion, and in 1 case postzygotic nondisjunction and postzygotic misdivision were the most likely mechanisms of formation. Alternative mechanisms are suggested in the remaining 2 cases.
Similar content being viewed by others
Introduction
The first clinical description of a child with a de novo supernumerary isochromosome 18p appeared in 1963 [1]; more than 50 cases have since been published. The tetrasomy 18p syndrome is characterized by psychomotor retardation, microcephaly, long facies with oval shape, high-arched eyebrows, short palpebral fissures and small pinched nose. Facial asymmetry is not unusual. Scoliosis and/or kyphosis, long fingers with contractures and feet malformations are frequent [1–3].
The chromosome 18 specificity of these marker chromosomes can now be confirmed by in situ hybridization with chromosome-18-specific probes [2, 4, 5]. Although previous reports determined only the chromosome 18 derivation of the marker, it can now be unambiguously shown using an 18p-specific library, that the marker chromosome is composed of only short-arm material [6].
Determination of the parental origin of the i(18p)s has previously been hampered by the lack of cytogenetic heteromorphisms of chromosome 18. Several models have been proposed to explain the formation of isochromosomes, but to our knowledge the mechanisms leading to tetrasomy 18p have not been clarified.
We have investigated 9 individuals with de novo tetrasomy 18p and their parents at polymorphic loci to determine the parental origin and mechanism of formation of the additional i(18p) chromosome. The cases were diagnosed by conventional chromosome banding and verified as i(18p)s by fluorescence in situ hybridization (FISH) using a flow-sorted 18p-specific library.
Material and Methods
Patient Population
The 9 patients were 4 girls and 5 boys, referred to cytogenetic analysis because of dysmorphic features and mental retardation. An overview of the patients is given in table 1.
Cytogenetic Studies
Chromosome analysis was performed on cultured lymphocytes using one or more of the following standard methods: QFQ-, CBG-, GTG-, and prometaphase RBA-banding as described [5–8].
Fluorescence in situ Hybridization
Fluorescence in situ hybridization (FISH) analysis was done using a biotin-dUTP-labeled chromosome 18 centromere-specific probe (ONCOR) according to the manufacturer’s instructions. In case 2, in situ hybridization had been performed using a radioactively labeled probe [5].
An 18p-specific library was constructed by flow sorting of an i(18p), and the flow-sorted material was amplified and biotinylated by degenerate oligonucleotide primed-polymerase chain reaction (DOP-PCR) [6]. Probe preparation, hybridization, detection and visualization were performed in all cases by one of us (E.B.) as previously described [6, 9]. An 18q-specific library was also used as control. The construction of this library, labeling, and FISH analysis were performed as above.
DNA Analysis
DNA was extracted from EDTA-anticoagulated blood by a salting-out procedure [10]. Eight DNA polymorphisms were detected after PCR amplification of genomic DNA [11]. End-labeling of primer, PCR amplification conditions, Polyacrylamide gel electrophoresis of the amplification product, and autoradiography were performed as described elsewhere [12]. The microsatellites used were (CA)n repeats at the following loci: D18S170 and D18S40 [13], D18S59, D18S63, D18S54, D18S62, D18S53, and D18S71 [14]. The scoring of polymorphic alleles was performed based on the knowledge of the karyotype showing tetrasomy 18p, i.e. an 18p microsatellite showing homozygosity for one allele in the proband would be interpreted as four copies of the same allele (1111), two alleles of almost equal intensity in the proband would be scored as 1122, two alleles with the smallest allele of much stronger intensity would be scored as 1112, and three alleles with the smallest allele of significantly stronger intensity would be scored as 1123, etc. The scoring was analogous to that of a trisomic condition, where dosage analysis by visual inspection was shown to correlate with densitometric reading of allele intensities [15].
Results
Parental Age Distribution
Range for mothers 24–39 years, mean maternal age 29 years; range for fathers 25–45 years, mean paternal age 32.1 years (table 1).
Chromosome analysis revealed 47 chromosomes in all 9 cases; the size and banding pattern of the extra small metacentric chromosome were compatible with an isochromosome of the short arm of chromosome 18. All cases were nonmosaic. All parental karyotypes were normal.
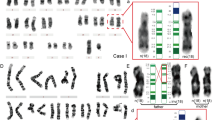
In situ hybridization with a chromosome 18 centromere-specific probe showed a monocentromeric signal on the marker chromosome in all cases. FISH using an 18p-specific library showed labeling of the short arms of chromosomes 18 and the whole of i(18p). No labeling of the marker chromosome was observed after FISH with an 18q-specific library (fig. 1).
DNA analysis indicated that all nine i(18p) chromosomes were of maternal origin (table 2, fig. 2). An uniparental origin of the normal chromosomes 18 could be excluded. In 6 cases (cases 1–6), the 18p markers showed that the maternal heterozygous alleles were reduced to homozygosity in the proband in the pericentromeric re-
Discussion
In their original report, published in 1963, before the banding era, Frøland et al. [1] suggested that the extra chromosome was an isochromosome of the short arms of chromosome 17 or 18. Later, with banding an isochromosome for 18p was suspected. However, by classical chromosome analysis it is sometimes difficult to distinguish an i(18p) from a deleted chromosome 18 [4, 6–8].
In the present cases, FISH analysis with a centromere-specific probe as well as with 18p- and 18q-specific libraries, showed that the marker chromosome was monocentric, and that it was unequivocally an i(18p), since the whole of the marker was painted with the 18p-specific library, with no labeling using the 18q library (fig. 1).
Formation of a nonmosaic de novo tetrasomy 18p due to an additional monocentric isochromosome 18p requires two events: nondisjunction and centromeric misdivision (= mitotic centromeric fission with fusion of sister p-arms).
The segregation of polymorphic DNA markers in different categories of premeiotic, meiotic or mitotic errors is presented in figure 3. The DNA markers used in this study cover the entire length of the short arm of chromosome 18 [16]. In 6 cases (cases 1–6), we found that the maternal heterozygous alleles were reduced to homozygosity in the pericentromeric region in the proband but retained heterozygosity in the telomeric region (table 2). To explain those 6 cases, we propose a model in which the tetrasomy is a result of meiosis II nondisjunction followed by postzygotic misdivision of one of the chromosomes with subsequent loss of all long-arm material (fig. 3, II).
Schematic representation of the segregation of a locus A near the telomere and a locus B near the centromere, each with two alleles (A, a and B, b) in normal meiosis and following premeiotic, meiotic, or postzygotic errors. Cross-over in meiosis I is indicated with ×. Not all the products of meiosis are shown. R = Reduction to homozygosity in the individual with tetrasomy 18p, given that the polymorphism was heterozygous in the parent who contributed the supernumerary isochromosome 18p; NR = nonreduction to homozygosity in the individual with tetrasomy 18p, given that the polymorphism was heterozygous in the parent who contributed the supernumerary isochromosome 18p; MI = meiosis I; MII = meiosis II; PZG = postzygotic division; NDJ = nondisjunction; MD = centromeric misdivision.
An alternative series of events compatible with the data is a premeiotic (mitotic) nondisjunction followed by a centromeric misdivision in MII [17].
In case 9, 6 of the 8 DNA markers were informative and showed reduction to homozygosity of the maternal alleles in the proband (table 2). Only postzygotic nondisjunction of chromosome 18 followed by misdivision of one of the chromosomes can explain these results (fig. 3, III). This assumes that no marker between the centromere and D18S40 shows nonreduction to homozygosity of maternal alleles. A postzygotic misdivision of one of the chromosomes in a maternally derived trisomy 18 may theoretically occur on the paternal chromosome in one third of the cases. Such cases would have maternal uniparental disomy for the long arm of chromosome 18. We tested this possibility with DNA markers mapping to the long arm of chromosome 18. At three informative 18q loci: D18S36, D18S35, and D18S38 biparental inheritance was demonstrated (data not shown). Usually mosaicism would be expected in cases of postzygotic nondisjunction. Analysis of 20 mitoses in blood and 59 mitoses in fibroblast cultures did not disclose any mitosis lacking the i(18p).
In 2 cases (cases 7 and 8), the probands were heterozygous for all informative maternal alleles. Unfortunately, the one or two markers, respectively, located closest to the centromere were not informative as the mothers were homozygous (table 2). If the maternal alleles in these probands were all nonreduced, the i(18p) formation could be explained by two previously postulated mechanisms: a centromeric misdivision occurring during a germinal premeiotic mitosis followed by MI nondisjunction [18] (fig. 3, IV), or an MI nondisjunction followed by a centrometric misdivision of one of the univalents during meiosis II [19] (fig. 3,I).
The proposed models for formation of the tetrasomy 18p are based on deduced haplotypes for the normal chromosomes 18 and the isochromosome 18p. However, in cases 7 and 8 alternative haplotypes could be determined from the pattern of the polymorphisms in the families. Determination of the i(l 8p) haplotypes, e.g. by PCR analysis of flow-sorted or microdissected i(18p) chromosomes would allow us to verify whether the deductions are correct or not.
Nondisjunction of chromosome 18 must be involved in the formation of tetrasomy 18p. DNA studies of trisomy 18 have shown that the majority of cases result from a maternal MII error [16]. This is in agreement with the most common model we propose, invoking MII nondisjunction as the first step in the formation of tetrasomy 18p.
Molecular analyses have also demonstrated that paternally derived trisomy 18 occurs in less than 5% of cases [16, 20]. In the present study, the isochromosome 18p was of maternal origin in all 9 cases.
Trisomy 18 is associated with advanced maternal age. Fisher et al. [16] found that the maternal age effect is significant in the MII error group. So far, an association of advanced parental age with tetrasomy 18p has not been demonstrated. In 1978, Nielsen et al. [7] found a mean maternal age of 30 years for the 25 cases of tetrasomy 18p reported previously. However, none of these papers explained the mechanisms involved in the i(18p) formation. Since we invoke that maternal MII nondisjunction is involved in several of the cases, increased maternal age would be expected. However, in the present limited number of cases, maternal age was increased in only 2 cases (cases 4 and 6, table 1).
The results of the present study serve to demonstrate that no single mechanism can account for all i(18p) cases. Based on genotyping, both premeiotic, meiotic and postzygotic division errors can be involved alone or in combination. Further studies are needed to determine the parental origin and to clarify the mechanisms of formation.
References
Frøland A, Holst G, Terslev E: Multiple anomalies associated with an extra small autosome. Cytogenetics 1963;2:99–106
Back E, Toder R, Voiculescu I, Wildberg A, Schempp W: De novo isochromosome 18p in two patients: Cytogenetic diagnosis and confirmation by chromosome painting. Clin Genet 1994;45:301–304
Schinzel A: *trip(18p)* = additional isochromosome 18p: in Schinzel A (ed): Catalogue of Unbalanced Chromosome Aberrations in Man. Berlin, Walter de Gruyter, 1984, pp 620–622.
Callen DF, Freemantie CJ, Rigenbergs ML, Baker E, Eyre HJ, Romain D, Haan EA: The isochromosome 18p syndrome: Confirmation of cytogenetic diagnosis in nine cases by in situ hybridization. Am J Hum Genet 1990;47:493–498
Blennow E, Nielsen KB: Molecular identification of a small supernumerary marker chromosome by in situ hybridization: Diagnosis of an isochromosome 18p with probe L1.84. Clin Genet 1991;39:429–433
Blennow E, Nielsen KB, Telenius H, Carter NP, Kristoffersson U, Holmberg E, Gillberg C, Nordenskjöld M: Fifty probands with extra structurally abnormal chromosomes characterized by fluorescence in situ hybridization. Am J Med Genet 1995;55:85–94
Nielsen KB, Dyggve H, Friedrich U, Hobolth N, Lyngbye T, Mikkelsen M: Small metacentric nonsatellited extra chromosome. Hum Genet 1978;44:59–69
Côté GB, Petmezaki S, Bastakis N: A gene for hypospadias in a child with presumed tetrasomy 18p. Am J Med Genet 1979;4:141–146
Blennow E, Telenius H, Larsson C, de Vos D, Bajalica S, Ponder BAJ, Nordenskjöld M: Complete characterization of a large marker chromosome by reverse and forward painting. Hum Genet 1992;90:371–374
Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A: A simple and efficient nonorganic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res 1989;17:8390.
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N: Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985;230:1350–1354
Petersen MB, Frantzen M, Antonarakis SE, Warren AC, Van Broeckhoven C, Chakravarti A, Cox TK: Comparative study of microsatellite and cytogenetic markers for detecting the origin of the nondisjoined chromosome 21 in Down syndrome. Am J Hum Genet 1992;51:516–525
Straub RE, Speer MC, Luo Y, Rojas K, Overhauser J, Ott J, Gilliam C: A microsatellite genetic linkage map of human chromosome 18. Genomics 1993;15:48–56
Gyapay G, Monssette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S: The 1993–94 Généthon human genetic linkage map. Nat Genet 1994;7:246–339
Petersen MB, Schinzel AA, Binkert F, Tranebjaerg L, Mikkelsen M, Collins FA, Economou EP: Use of short sequence repeat DNA polymorphisms after PCR amplification to detect the parental origin of the additional chromosome 21 in Down syndrome. Am J Hum Genet 1991;48:65–71
Fisher JM, Harvey JF, Morton NE, Jacobs PA: Trisomy 18: Studies of the parent and cell division of origin and the effect of aberrant recombination on nondisjunction. Am J Hum Genet 1995;56:669–675
Vig BK: Sequence of centromere separation another mechanism for the origin of nondisjunction. Hum Genet 1984;66:239–243
Rivera H, Rivas F, Cantú JM: On the origin of extra isochromosomes. Clin Genet 1986;29:540–541
Van Dyke DL, Babu VR, Weiss L: Parental age, and how extra isochromosomes (secondary trisomy) arise. Clin Genet 1987;32:75–80
Bugge M, Petersen MB, Brandt CA, Hertz JM, Mikkelsen M: DNA studies in trisomy 18. Report of the Second International Workshop on Human Chromosome 18 Mapping 1993. Cytogenet Cell Genet 1994;65:141–165
Acknowledgements
We wish to thank photographer Jette Bune Rasmussen for the layout of the figures. This study was supported by the Danish Biotechnology Research and Development Programme 1991–1995, grant 16/92 from Fonden til Laegevidenskabens Fremme, Kong Christian den Tiendes Fond, Direktoer Jacob Madsens og Hustru Olga Madsens Fond, and Broedrene Hartmanns Fond.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bugge, M., Blennow, E., Friedrich, U. et al. Tetrasomy 18p de novo: Parental Origin and Different Mechanisms of Formation. Eur J Hum Genet 4, 160–167 (1996). https://doi.org/10.1159/000472190
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472190
Key Words
This article is cited by
-
Prenatal genetic diagnosis of tetrasomy 18p from maternal trisomy 18p: a case report
Molecular Cytogenetics (2022)
-
Pallister-Killian syndrome: clinical, cytogenetic and molecular findings in 15 cases
Molecular Cytogenetics (2018)
-
Adults with Chromosome 18 Abnormalities
Journal of Genetic Counseling (2015)
-
Tetrasomie 18p
Monatsschrift Kinderheilkunde (2008)
-
De novo isochromosome 18p in a female dysmorphic child
Journal of Applied Genetics (2006)