Abstract
Wildlife populations are under intense anthropogenic pressures, with the geographic range of many species shrinking, dramatic reductions in population numbers and undisturbed habitats, and biodiversity loss. It is postulated that we are in the midst of a sixth (Anthropocene) mass extinction event, the first to be induced by human activity. Further, threatening vulnerable species is the increased rate of emerging diseases, another consequence of anthropogenic activities. Innovative approaches are required to help maintain healthy populations until the chronic underlying causes of these issues can be addressed. Fibropapillomatosis in sea turtles is one such wildlife disease. Here, we applied precision-medicine-based approaches to profile fibropapillomatosis tumors to better understand their biology, identify novel therapeutics, and gain insights into viral and environmental triggers for fibropapillomatosis. We show that fibropapillomatosis tumors share genetic vulnerabilities with human cancer types, revealing that they are amenable to treatment with human anti-cancer therapeutics.
Similar content being viewed by others
Introduction
Wild sea turtle populations, which are already in danger of extinction (http://www.iucnredlist.org/), are under increasing threat from a potentially fatal virulent tumor, fibropapillomatosis (Fig. 1a). These tumors are undermining turtle conservation efforts within Florida (FL, where fibropapillomatosis was first recorded) and circum-globally, with the disease having spread throughout equatorial and subequatorial equatorial regions1,2,3,4,5,6,7,8,9. Green sea turtles (Chelonia mydas) are the species most commonly and severely afflicted with fibropapillomatosis (Fig. 1), but it also occurs in all other sea turtle species4,5,6,7,10,11,12, and evidence suggests that its geographic range is spreading6,8,9,13. Fibropapillomatosis is now being reported from more northern latitudes where it has never been previously recorded6. The etiology of fibropapillomatosis is not fully understood, but the disease has a combination of viral and environmental cofactors5. A large number of oncogenic viruses have been reported in terrestrial and aquatic wildlife and domestic animals14,15. Meanwhile, in humans, the number of cancer incidences identified as attributable to pathogens has tripled in only 4 years16,17, now accounting for 15% of all human cancers. Furthermore, emerging infectious disease events in humans are dominated by zoonoses (diseases transmitted to humans from animals, 60.3%), with the majority of these (71.8%) originating in wildlife18,19,20. Many emerging infectious diseases (25–44%) are viral or prion in nature18,19,21,22. It is clear that vertebrate virus-borne disease can spread through the marine environment, and that anthropogenic effects (e.g., habitat degradation) are largely responsible for the increased rate in the emergence of infectious diseases4,18,23,24,25. Therefore, a more concerted effort to improve our understanding of pathogen-induced cancers in wildlife, including potential exacerbating effects of human coastal land use, is crucial for enhancing both human and wildlife health.
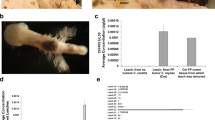
Fibropapillomatosis tumors and differential transcript expression. a Fibropapillomatosis-afflicted green sea turtles (Chelonia mydas). Left and center: Turtles at the Whitney Sea Turtle Hospital, FL, prior to tumor removal surgery. Right: High magnification image of a fibropapilloma tumor, which was profiled by RNA-seq. b Principle component analysis of patient-matched control and fibropapillomatosis RNA-seq samples. Upon admittance to the hospital, Swoope (sp) had a tumor score of class 3 (Hawaii classification system)105, and severe (FPI > 185.5) (Southwest Atlantic classification system)106, while Major (mj) had a tumor score of class 2105, and moderate (FPI = 81.5)106. c Overlap of transcripts differentially expressed in fibropapillomatosis tumors, independently called by DESeq2 and EdgeR from the RNA-seq data. Area-proportional Venn diagrams were generated using BioVenn (http://www.biovenn.nl/)107. Transcripts were considered to be DE if passing the following cut-offs: false discovery rate (EdgeR) or adjusted p-value (DESeq2) of <0.05 and log2 fold change of >2 or <−2
Despite fibropapillomatosis first being reported in the late 1800s/early 1900s, and having reached panzootic levels in the 1990s1,2,3,5,6,26, practically nothing is known about the host molecular events/genes driving the tumors. Furthermore, the viral dynamics of fibropapillomatosis are poorly understood. Transmissibility of the tumors was experimentally proven in 199527. Fibropapilloma tumors are transmissible to fibropapillomatosis-free turtles via injection of tumor-derived freeze–thawed and filtered sub-cellular fractionations, indicating a viral agent27. The inability to culture the responsible virus in a lab setting28,29 has meant that Koch’s postulates remain unfulfilled and viral causation has yet to be definitively proven. However, a number of studies have shown fibropapilloma-associated turtle herpesvirus (FPTHV), also known as chelonid herpesvirus 5 (ChHV5) and chelonid fibropapilloma-associated herpesvirus (CFPHV), to be associated with fibropapillomatosis globally5,10,27,30,31, and the genome of this virus has been partially sequenced32,33. Conversely, high levels of ChHV5 DNA have been detected in clinically healthy marine turtles30,34. Recently a step toward completing Koch’s postulates was made with the successful in vitro growth of ChHV5, in tumor cells cultured in a replica complex three-dimensional structure of turtle skin, including both fibropapilloma tumor fibroblasts and turtle keratinocytes35.
ChHV5 appears to be a virus upon which oncogenic potential is conferred by a form of external environmental perturbation. This is indicated by the widespread presence of ChHV5 in clinically healthy turtles, including in regions where fibropapillomatosis has never been observed (e.g., Europe) and in non-tumor tissues of fibropapillomatosis-afflicted turtles7,30,34,36,37. Furthermore, phylogenetic reconstruction analyses have shown that ChHV5 has been present in sea turtles well before the outbreak of fibropapillomatosis, having co-evolved with its turtle hosts5,36,38,39,40. Current estimates for the virus diverging to becoming specific to marine turtles range from hundreds to millions of years, yet tumors are thought to be only a relatively recent phenomenon5,36,38,39,40. Strong correlative evidence suggests that localized human-induced environmental changes are responsible for conferring oncogenic potential upon ChHV55,41,42,43,44. Fibropapillomatosis rates are extremely high in many habitats degraded by human activity, yet almost absent from neighboring more pristine areas5,43,44.
Currently, surgical removal is the primary treatment for turtles with fibropapillomatosis, but one study showed that almost 60% of tumors (in 38.5% of cases) regrew post-surgery7. The regrowth occurs within an average of 36 days, but since records are only available for the relatively short period of captive rehabilitation, it is likely that the true occurrence of regrowth is much higher. Additionally, there is currently no effective treatment for turtles presenting with internal tumors (which can result in organ failure), detectable by X-ray, magnetic resonance imaging (MRI), or computerized axial tomography (CAT) scanning, with such turtles usually having to be euthanized. Therefore, novel therapies are urgently needed to enhance the rehabilitation outcomes of turtles with fibropapillomatosis.
Although the viral nature of fibropapilloma tumors has been widely, if not conclusively, investigated, the role of the host molecular events in driving these tumors has been largely overlooked. Given that oncogenic viruses play their most critical role in the very early stages of tumor development, primarily in the transformation of host cells, the scope of anti-viral treatments for tackling well-established tumors is severely limited45. This fact is highlighted by the hit-and-run hypothesis of viral oncogenic transformation, whereby after inducing the malignant conversion of host cells, viruses are no longer necessary for the maintenance of the malignant state and may no longer be physically present in tumors46. Therefore to improve our understanding and treatment of fibropapillomatosis, it is particularly important to examine the molecular mechanisms present in host cells, which drive the growth of established tumors. That turtles tend to present to rehabilitation facilities in the more advanced stages of tumor growth highlights the critical knowledge gap in our understanding of fibropapillomatosis.
Here, we demonstrate the applicability of precision wildlife medicine4 to improving animal health and advancing wild animal clinical care. In order to determine the molecular signaling events responsible for driving fibropapilloma tumor growth in green sea turtles (C. mydas), we employed precision medicine approaches47,48,49, previously honed in the field of human oncology. This included next-generation Illumina RNA sequencing (RNA-seq) transcriptomic profiling and computationally-based systems-level analysis. We show that in established tumors, there is minimal ChHV5 expression; rather tumors are primarily driven by altered host gene expression, leaving them vulnerable to anti-cancer therapeutics. We thereby advanced the study of host molecular drivers of chelonid fibropapilloma tumors from their current pre-genetic era, bypassing the candidate gene-centric approach straight into the modern genomics era, providing a solid understanding of the basic cell signaling pathways that drive tumor growth, thus enabling the identification of therapeutic targets for fibropapillomatosis treatment.
Results
Fibropapillomatosis transcriptomics reveals altered host gene expression
We transcriptomically profiled tumor tissue, surgically removed by laser resection as part of the turtle’s rehabilitative care, and compared global gene expression to that of biopsies from non-tumor areas of the same turtles. Ten RNA samples from two juvenile green turtles (C. mydas), which had stranded in northern FL were used for sequencing (Fig. 1b). The early development phase of fibropapilloma tumors is associated with proliferation of epidermal cells, while later growth is associated with proliferation of the dermal layer5. Therefore, in order to obtain representative untransformed control tissue, we profiled skin punch biopsies containing epidermis, dermis, and subcutaneous tissue. Principle component analysis of the RNA-seq data confirmed that non-tumor samples grouped separately from tumor samples (Fig. 1b). A de novo transcriptome was assembled from the sequencing reads using Trinity v2.1.150. The transcriptome was generated using Trinity’s unguided function, to ensure that any tumor-specific or viral transcripts present would not be filtered out during transcriptome generation, and given that currently only a draft C. mydas genome exists. The de novo transcriptome consisted of 266,069 transcripts, with an N50 length (the largest length such that 50% of all base pairs are contained in contigs of this length or larger) of 1562 bp. Pair-wise comparison of the tumored and non-tumored tissue revealed that 4345 transcripts were differentially expressed (DESeq2, adjusted p-value cut off of 0.05 and with a log2 ± 2 fold change applied) (Fig. 1c), with the majority of these (83%) being upregulated in the tumors (Supplementary Figure 1A, B). When EdgeR was used to independently call differentially expressed (DE) transcripts, there was a strong overlap between the transcripts called as DE between DESeq2 and EdgeR (Fig. 1c), adding confidence that these are genuine DE transcripts and not artifacts of the computational analysis. We confirmed the upregulation of Fibronectin Type III Domain Containing 1 (FNDC1, a cancer-associated gene51), a top DE transcript (Fig. 2a), in an extended panel of fibropapillomatosis tumor samples (14 control and 67 tumor samples) by qPCR (Fig. 2b).
Differentially expressed host (C. mydas) and viral (ChHV5) transcripts. a Top ten statistically significant upregulated and top ten downregulated transcripts in fibropapillomatosis tumors compared with control tissue. Transcript expression values underlying the differential expression analysis are provided in Supplementary Table 4. Differential expression p-values reported are adjusted p-values, generated by DESeq2 by a Wald test followed by Benjamini–Hochberg correction. b Relative expression of Fndc1 mRNA in fibropapillomatosis tumor and control samples, as detected by RT-qPCR. Data for 66 tumor samples and 14 control samples. The p-value was calculated by t-test. c Expression levels (trimmed mean of M values, TMM) of ChHV5 viral RNA de novo transcripts across the ten RNA-seq samples. Related transcript variants are denoted by t.1, t.2, or t.3
RNA-seq reveals a paucity of ChHV5 viral transcripts within tumors
Having confirmed the differential expression of host genes within the tumors, we next examined ChHV5 viral expression by blasting the de novo transcriptome against the ChHV5 partial genome33 [GenBank accession number: HQ878327.2]. Although ChHV5 has been associated with fibropapillomatosis and ChHV5 DNA is readily detectable within tumors5,31, there was an almost complete lack of viral transcripts within the RNA-seq samples, with only four ChHV5 transcripts being detected, three of which were related transcript variants (Fig. 2c, Supplementary Table 1). Interestingly, no viral transcripts were detected in the non-tumor tissue samples, being specific to the tumor samples (Fig. 2c). However, even in the tumors, there was a paucity of viral transcripts expressed, with the transcripts related to only two regions of the genome; the F-LANA/HP2 and the HP3/HP4 regions (Fig. 2c). Expression levels of these transcripts were also relatively low (viral expression range: 0–1.344 trimmed mean of M values (TMM). The range of expression for all transcripts [viral and host] was 0–125,636.734 TMM, with the average expression level of all transcripts across samples being 4.608 TMM.) Within the ChHV5 genome, the coding region of the F-LANA and HP2 genes overlap, one transcript from the RNA-seq mapped to this region (Fig. 2c and Supplementary Table 1). However, for the HP3/HP4 region, three related but distinct transcript variants were called (Supplementary Table 1), one within the HP3 gene and two mapping to the region between the HP3 and HP4 genes (Fig. 2c and Supplementary Table 1). Due to a genomic duplication event33, all four viral transcripts detected map to two locations within the ChHV5 genome, at both the terminal and internal repeat regions (Supplementary Table 1).
To confirm the paucity of viral transcripts independently of our de novo transcriptome, we also aligned the reads for each sample against the ChHV5 genome. As with the de novo transcriptome analysis, direct mapping to the ChHV5 genome also showed viral reads predominantly aligning to the terminal and internal repeat region around the F-LANA, HP2, HP3, and HP4 genes (Supplementary Figure 1C). F-LANA is a latency-associated nuclear antigen gene, which promotes viral latency by suppressing viral transcription and facilitating DNA replication52. The detection of the F-LANA expression along with the paucity of other ChHV5 transcripts suggests that ChHV5 is latent within these tumors. Viral latency would indicate that any causative role for ChHV5 in fibropapillomatosis tumorigenesis occurs early, and that the virus does not continue to drive the growth of established tumors.
Fibropapillomatosis has a neuronal signature and similarity to human cancer
To determine the molecular mechanisms driving fibropapillomatosis tumors, we took the top 600 (300 per direction of regulation, Supplementary Table 2) statistically significant DE transcripts (DESeq2 analysis, adjusted p-value cut-off of 0.05 and with a log2 ± 2 fold change applied, the adjusted p-value ranges of the 300 upregulated and downregulated transcripts were 3.52E−119 to 3.15E−14 and 1.33E−105 to 3.35E−05, respectively), determined their closest characterized homolog using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi)53, and performed systems level analysis. Network analysis of these transcripts (STRING database v10.5, http://www.stringdb.org/)54 revealed a highly interconnected network, which was strongly enriched for neuronal-related genes (Fig. 3a, b), with nervous system development and neurogenesis gene ontology (GO) terms being strongly enriched (FDR padj. = 2.89E−12 and 2.03E−11, respectively).
Transcripts differentially expressed in fibropapillomatosis are associated with nervous system development. a Protein–protein interaction map of the top differentially expressed transcripts (RNA-seq) with homology to characterized human genes. Generated from the top 600 (300 per direction of regulation) differentially expressed transcripts. Nervous system development-related nodes are highlighted by red shading. Network generated by the STRING database54 (v10.5, http://www.stringdb.org), with the inbuilt KEGG pathway enrichment analysis tool applied to this network. b Fold change in expression of the differentially expressed transcripts (RNA-seq) related to nervous system development (KEGG), listed in order of false discovery rate (FDR, KEGG pathway enrichment analysis tool). Transcript expression values underlying the differential expression analysis are provided in Supplementary Table 4
We next performed Ingenuity Pathway Analysis (IPA, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/)55, which revealed that fibropapilloma tumors share a striking similarity to a number of human cancers (Fig. 4a–c). Human basal cell carcinoma (BCC) signaling was the top-ranked signaling pathway in fibropapillomatosis tumors when compared with non-tumored control tissue (p = 1.77E−10, Fig. 4a), with BCC signaling being activated (Fig. 4b). Mirroring the network analysis findings, axonal guidance signaling was the second most significant pathway identified (p-value = 2.45E−10) (Fig. 4a). In addition to their role in neuronal development and neuronally-derived tumors (e.g., medulloblastoma and neuroblastoma), axonal guidance genes have been shown to be involved in driving tumorigenesis in a range of other non-neuronally-derived human cancer types such as pancreatic cancer56. Human embryonic stem cell pluripotency signaling was the third most highly ranked pathway (p = 2.34E−08), in line with the fibropapillomatosis tumors consisting of less differentiated more stem-like cells, as is the case in human tumors57,58.
Pathway, disease gene ontology (GO) term, and transcriptional regulator analysis of the top 600 transcripts differentially expressed in fibropapillomatosis tumors. a Top 50 canonical pathways of the fibropapillomatosis tumor differentially expressed transcripts (RNA-seq), as detected by IPA, ranked by p-value (calculated by right-tailed Fisher’s Exact Test, with Benjamini–Hochberg correction). b Activation/inhibition z-scores of the canonical pathways of the fibropapillomatosis tumor differentially expressed transcripts, as detected by IPA. c Activation z-scores of the top 25 disease-associated GO terms of the fibropapillomatosis tumor differentially expressed transcripts (RNA-seq), as detected by IPA, ranked by p-value (calculated by right-tailed Fisher’s Exact Test, with Benjamini–Hochberg correction). d Activation/inhibition z-scores of the inferred transcriptional regulators (ITRs) of the fibropapillomatosis tumor differentially expressed transcripts (RNA-seq), as detected by IPA, ranked by p-value (calculated by right-tailed Fisher’s Exact Test, with Benjamini–Hochberg correction). Legend of x-axis labels for Fig. 3a–c is also provided in Supplementary Table 5
Cancer and neuronal disease terms also topped the disease GO terms analysis (Fig. 4c and Supplementary Figure 1C), with cancer growth, invasion, and metastasis terms being activated, while cell death and apoptosis terms were inhibited (Supplementary Figure 1C), again consistent with fibropapillomatosis tumors tending to have rapid growth rates, and a propensity for tens to hundreds of tumors to develop on a single infected individual. Strikingly, of the 334 unique genes (from the top 600 DE transcripts) with homology to human genes, almost all of them (321 genes) are associated with human cancer (Fig. 4c, p-value = 3.10E−06). This includes the upregulation of genes such as ITGA5 (Supplementary Table 2), which are involved in virally-mediated angiogenesis promotion in Kaposi’s sarcoma59.
IPA inferred transcriptional regulator (ITR) analysis, which matches observed DE genes to known patterns of gene regulation by specific transcriptional regulators55, revealed a highly interconnected network of regulators, governing the gene expression changes seen in fibropapillomatosis (Supplementary Figure 1D). The ITR analysis also suggested a number of potential novel therapeutics for fibropapillomatosis treatment such as β-estradiol, or inhibitors of a number of pathways including TGFβ, Wnt, MAPK, and EGFR (Fig. 4d).
Wnt and SHH signaling regulate fibropapillomatosis, which correlates with UV exposure
Our systems-level analysis identified shared molecular networks between sea turtle fibropapillomatosis and human BCC, including Wnt, BMP, and SHH signaling. While being involved in a variety of other cancer types60,61,62,63,64,65,66, Wnt and sonic hedgehog (SHH) signaling are a part of a shared regulatory network in BCC and are differentially activated in fibropapilloma tumors (Fig. 5a, b). We confirmed that Wnt5a was differentially expressed in a large panel of fibropapilloma tumors, primarily being upregulated (Fig. 5c). Wnt5a is an important regulator of both the Wnt/PCP and Wnt/β-catenin pathways, playing a prominent role in neuronal stem cells and nervous system development67,68, and is an important regulator of tumorigenicity of many human cancers, including neuronal derived ones62,67,68,69,70,71,72.
Wnt, SHH, and BMP pathways in fibropapillomatosis tumors. a Left: Activation/inhibition z-scores of Wnt- and SHH-related IPA pathway analysis findings of the fibropapillomatosis tumor differentially expressed transcripts (RNA-seq), p-values calculated by right-tailed Fisher’s Exact Test, with Benjamini–Hochberg correction. Right: Interaction map of the 43 Wnt, SHH, and BMP pathway component and target genes differentially expressed in fibropapillomatosis tumors (RNA-seq). Image generated using GeneMANIA (v3.5.0, http://genemania.org/)104. In addition to the 43 DE genes (nodes with stripped shading), 20 closely functionally-related network components are also shown (nodes with uniform shading). Basal Cell Carcinoma KEGG pathway score of the DE components of the network is shown below the network. b Fold change in expression of the differentially expressed transcripts (RNA-seq) of Wnt, SHH, and BMP pathway-related genes. When more than one transcript of a gene was present in the top 600 DE genes (300 per direction), each transcript is denoted by t1, t2, t3, etc. Differential expression p-values reported are adjusted p-values, generated by DESeq2, by a Wald test followed by Benjamini–Hochberg correction. Transcript expression values underlying the differential expression analysis are provided in Supplementary Table 4. c Relative expression of Wnt5a mRNA in a panel fibropapillomatosis tumor and control samples, as detected by RT-qPCR. Data for 66 tumor samples and 13 control samples. The p-value was calculated by t-test
As BCC is a skin cancer with both genetics and ultraviolet radiation (UV) exposure as risk factors, we investigated whether any correlation between fibropapillomatosis incidence and high UV exposure levels exists. Historical daily UV index (UVI) data, dating from 1995 to 2015, were available for three coastal cities in FL: Jacksonville (north FL), Tampa Bay (central FL), and Miami (south FL) (http://www.ftp.cpc.ncep.noaa.gov/long/uv/cities/ and http://www.cpc.ncep.noaa.gov/products/stratosphere/uv_index/uv_annual.shtml). Despite fibropapillomatosis’s rapid circum-global spread (including the Caribbean, Hawaii, Australasia, India, and Africa), fibropapillomatosis has been much slower to spread to more temperate latitudes, with northern FL (north of latitude 29°N) remaining fibropapillomatosis-free until 2000, despite fibropapillomatosis being present in the FL Keys since before 19131,2,3,6. Therefore, while fibropapillomatosis was present in the Miami and Tampa areas for the entire duration for which UVI recordings are available, the Jacksonville data cover the period of the spread of fibropapillomatosis into northern FL (Fig. 6a). There was a clear trend from north to south in the number of days annually with extreme UV exposure levels (UVI > 11), and the number of extreme UVI days has increased from 1995 to 2015 across all three FL sites. Between 1995 and 2015, both the total number of green sea turtles (C. mydas) stranding in FL exhibiting fibropapillomatosis and the percentage of all stranded C. mydas in FL, which are afflicted with fibropapillomatosis has grown73 (Fig. 6b,c). The increasing incidence of fibropapillomatosis in FL is in line with global fibropapillomatosis incidence trends6. We next performed a linear regression analysis to determine whether there was any correlation between the number of C. mydas stranding with fibropapillomatosis each year and the number of days of extreme UVI (average of all three sites) in the previous year (Fig. 6d). UVI levels from the previous year were used due to the recognized lag between UV exposure and skin cancer development74. The analysis revealed that there was a mild positive correlation between UVI exposure and the numbers of stranded fibropapillomatosis-afflicted turtles (R2 = 0.493), suggesting that UV exposure may also be a risk factor for fibropapillomatosis and a potential limiting factor accounting for the relatively slow northward spread of fibropapillomatosis. The observed level of correlation is relatively high, considering that fibropapillomatosis is a multifactorial disease with viral and other environmental factors likely contributing to disease progression5.
Putative fibropapillomatosis prevalence and ultraviolet (UV) radiation exposure link. a Number of days annually in the extreme UV index (UVI) category recorded in Miami, Tampa Bay, Houston, and Jacksonville (data obtained from: http://www.ftp.cpc.ncep.noaa.gov/long/uv/cities/). Dates for the occurrence of fibropapillomatosis in the vicinity of each UVI sampling site obtained from Hargrove et al.6. b Annual percentage of green sea turtles stranding in FL afflicted with fibropapillomatosis, obtained from Foley et al.73. c Yearly averages of Jacksonville, Miami, and Tampa Bay UVI extreme days annually, vs. annual fibropapillomatosis-afflicted green turtle strandings in FL. Correlation between fibropapillomatosis strandings and UVI conducted using R2 linear regression analysis
5-FU post-surgical treatment mitigates fibropapillomatosis tumor regrowth
We revealed that fibropapillomatosis shares molecular characteristics with human BCC, for which a number of effective therapies exist including the topical application of fluorouracil (5-FU) onto affected skin regions. While 5-FU is used in a number of human cancers, topical application is a first line treatment in BCC75. Sixty percent of fibropapillomatosis tumors regrow after surgical removal7, with surgery being the only widely employed treatment option. As is the case in human oncology, adjunct drug/chemotherapy treatment post-surgery is likely to dramatically reduce tumor regrowth rates, thus the utility of 5-FU as an adjunct therapy in fibropapillomatosis was assessed in eye tumors.
Fibropapillomatosis tumors commonly occur in the eye region, often arising from the conjunctivae or cornea5,7 (Fig. 7a). Eye tumors can grow to a considerable size, obscuring vision. Given the smaller surface area of eye tumors and the high level of debilitation caused by them, these tumors are particularly suited to topically delivered post-surgery chemotherapy to help prevent tumor regrowth. Sixty-seven percent of turtles whose eye tumors (ocular and periocular) were treated with surgery-only experienced eye tumor regrowth (Fig. 7b, Supplementary Table 3), this level of regrowth is in line with the rates seen across all fibropapillomatosis tumors (regardless of body location) in other studies, i.e., 60% of tumors re-growing7. However, when turtles received 5-FU post-surgery (8 week 5-FU course, treated twice daily with 1% topical 5-FU drops), only 18% of turtles experienced eye tumor recurrence (Fig. 7b). The duration of post-treatment follow-up varied as dictated by the individual circumstance of each turtle (Supplementary Table 3), but was between 2 and 12 months.
Fibropapillomatosis eye tumor regrowth rates in the presence and absence of adjunct 5FU treatment. a Green sea turtles afflicted with fibropapillomatosis eye tumors. b Fibropapilloma eye tumor regrowth rates, when treatment consists of surgical removal only, or surgical removal followed by 8 weeks of topical 5-FU treatment. Total n = 121 turtles, with the number of turtles per condition inserted within the relevant section of each bar
Discussion
The increased rate of emerging diseases, and their exacerbating negative effects on already stressed wild populations, poses an additional burden on already elevated rates of population reduction and species extinction4,18,23,24,25,76,77,78,79,80,81. The application of omic technologies and human precision medicine approaches can rapidly help alleviate the burden of such diseases and assist in identifying causative factors, ultimately improving species conservation1. To this end, we applied next-generation transcriptomic profiling and systems level analysis to fibropapillomatosis tumors, revealing evolutionarily conserved molecular mechanisms between fibropapillomatosis and human cancer types such as BCC and neuronally-derived tumors. BCC itself is known to have neuronal links, including the SHH pathway and correlation between different BCC sub-types and neuronal differentiation markers82. The neuronal signature of fibropapillomatosis tumors suggests either extensive innervation in these tumors, a previously unappreciated role of neuronal blast cells in the origin of fibropapillomatosis tumors, or at least the involvement of neuronal-related genes in fibropapillomatosis growth. ChHV5 has previously been shown to reside in sea turtle neuronal cells37, supporting a neuronal origin hypothesis. To mitigate against any potential cell type-dependent gene expression biases and confirm our findings, our fibropapillomatosis sequencing data should be compared against additional C. mydas tissue-specific transcriptomic profiles as they become available.
Interestingly, we detected a paucity of ChHV5 transcripts in our sequencing data. While, this is in line with suggestions that ChHV5 remains latent in established tumors5,37 and is therefore largely transcriptionally inactive, analysis of a wider variety of fibropapillomatosis tumor transcriptomes is required to confirm this finding. Our tumor profiling also suggests a number of potentially promising novel therapeutic approaches. Adjunct 5-FU treatment dramatically reduced the post-surgery regrowth rate of eye tumors, demonstrating potential to reduce rehabilitation costs and burdens on individual turtles by negating the need for additional rounds of surgery, and shortening the rehabilitation period. The ability of 5-FU to successfully reduce fibropapillomatosis tumor recurrence rates reinforces the power of genomics-era analysis to rapidly identify novel therapeutic options for fibropapillomatosis treatment. While, 5-FU proved effective for adjunct fibropapillomatosis eye tumor therapy, it warrants further investigation in an independent, controlled clinical study. Such a study should also investigate 5-FU as a treatment for tumors in other body locations. However, as 5-FU is a cytotoxic drug, and in BCC, it is only effective against low-grade tumors75, more target therapies should also be trialed. Our analysis also predicts a number of pathways for which more targeted therapies exist, which are likely to have applicability as both adjunct and first-line treatments. SHH and Wnt signaling form an interconnected regulatory node in fibropapillomatosis tumors, the targeting of which could potentially prove effective for fibropapillomatosis treatment. Vismodegib (trade name, erivedge), an inhibitor of the SHH receptor Smoothened (SMO), is already in use clinically for the treatment of BCC83, while a number of Wnt modulating therapies are being actively pursued as anti-cancer treatments, currently at the clinical trial stage84. Other targets include MAPK, TGFβ, and estrogen signaling. These are already used in the treatment of a number of human cancers, including neuronally-derived tumors, and their effectiveness as fibropapillomatosis therapeutics should be further investigated85,86,87,88,89,90. To fully achieve the promise of precision medicine and move toward more targeted treatment, newer generation therapeutics highlighted by our findings, such as vismodegib, should be trialed as anti- fibropapillomatosis treatments.
The shared molecular drivers between fibropapillomatosis and BCC suggested a putative role of UV radiation as one of the environmental co-triggers of fibropapillomatosis. Interestingly, Wnt and Metalloprotease signaling are two of the top DE pathways in fibropapillomatosis tumors. These pathways have been shown to be regulated by UV exposure in human skin cells and canine cornea, and mediate UV-induced abnormal morphogenesis in Hydra91,92,93,94. UV exposure has also been linked to systemic immunosuppression95. Immunosuppression also occurs in turtles with fibropapillomatosis, although it has not yet been proven if this is a cause or a consequence of fibropapillomatosis5. Taken together, further investigation into interactions between UV exposure, regulation of host gene expression including immune-related genes, and ChHV5 viral load are highly warranted. The correlation between fibropapillomatosis incidence and extreme UV conditions suggests that UV may be another risk factor for fibropapillomatosis. Fibropapillomatosis is most prevalent during green sea turtles’ juvenile inshore life-stage when they inhabit more UV-exposed waters5,10. Even in an artificial transmission setting, fibropapillomatosis tumors arose simultaneously, regardless of the time of initial inoculation, only after temperatures increased27. The authors attributed this to potential effects of water temperature27, though this seasonality is also a proxy for UV exposure. While our analysis focused on FL, UV exposure is also altering globally as a result of climate change96. In the case of fibropapillomatosis, UV exposure could be directly involved in oncogenic transformation, as is the case for human skin cancers, or alternatively could be acting through indirect mechanisms. For instance, increased UV could contribute to immunosuppression resulting in reduced control of ChHV5 infections95, increased frequency of algal bloom events, which have been correlated with fibropapillomatosis occurrence5,42,43,44,97, or UV radiation, which can alter the chemical composition of contaminants in inshore waters into carcinogenic forms98,99. Future research is required to determine whether UV exposure is causally linked to fibropapillomatosis tumor development, and if so, via which mechanism. The genomic profiling reported here has improved our molecular understanding of fibropapillomatosis by revealing the host signaling pathways involved in tumors and indicating novel rehabilitation strategies for fibropapillomatosis-afflicted sea turtles. Additionally, this study demonstrates the power of precision medicine approaches47,48 to tackle rare or understudied diseases for which there is limited background genetic knowledge, and provides a proof-of-principle study for the emerging field of precision wildlife medicine4.
Methods
Tissue sampling
Sampling was carried out under permit number MTP-17–236 from the Florida Fish and Wildlife Conservation Commission and with ethical approval from the University of Florida Institutional Animal Care and Use Committee (IACUC). Fibropapilloma tumors were surgically removed by laser resection as part of the turtle’s rehabilitative care. All samples were obtained from juvenile C. mydas, as this is the stage most afflicted by fibropapillomatosis5,10. As juveniles, the sex of an individual is not readily determinable. However, one of the turtles whose tumors were sequenced, Swoope (sp, patient ID: 02-2015-Cm), was later determined to be male during necropsy, after being euthanized due to the presence of internal tumors. Global gene expression of the tumors was compared with that of non-tumored areas of the same turtles, obtained by 4 mm punch biopsies during the tumor removal surgery. Non-tumored sites were selected by gross examination of the region by the attending veterinarian and confirmed visually to be tumor-free normal skin regions and not bordering any tumorous regions by the attending vet technicians and researchers. Samples were stored in RNA-later (Qiagen) at −80 °C, according to manufacturer’s instructions, for up to 3 months.
RNA extraction and RNA-seq
Total RNA was extracted using an RNeasy Kit (Qiagen) according to manufacturer’s instructions. DNA was digested with DNA-free Kit (Applied Biosystems). RNA quality was checked by RT-qPCR (as below) and on a 2100 Bioanalyser (Agilent) using a Eukaryote Total RNA Nano Chip, samples’ RIN value range 8.1–9.8. Ten RNA samples, comprising seven fibropapillomatosis tumor samples (shoulder and neck cutaneous verrucous fibropapilloma tumors) and three non-tumor samples, from two juvenile green turtles (C. mydas), which had stranded in Northern FL were used for sequencing (Fig. 1b). Sequencing libraries were generated from 500 ng of total RNA using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs), including ployA selection, according to manufacturer’s protocol. Size and purity of the libraries were analyzed on a Bioanalyser High Sensitivity DNA chip (Agilent). Libraries were paired-end sequenced with a read length of 75 bp a HiSeq 3000 (Illumina). ERCC Spike-In Mix (ThermoFisher) was used as an internal control, 2 µL of 1:400 diluted ERCC Spike-In Mix with 500 ng of total RNA input.
Reads were trimmed with Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove low-quality ends from reads in addition to adapter removal. A de novo transcriptome was constructed from the sequencing data using Trinity v2.1.150, specifying a minimum contig length of 300. Transcript abundance was quantified with RSEM using bowtie2 for the alignment using the align_and_estimate_abundance.pl script available through the Trinity toolkit. A matrix of gene counts (genes.counts.matrix) and a matrix of TMM-normalized expression values (TMM.EXPR.matrix) was constructed in RSEM using the abundance_estimates_to_matrix.pl script available through the Trinity toolkit100,101. A PCA plot (Fig. 1b) was generated using the PtR script in the Trinity toolkit.
Prior to differential expression analysis, the genes.counts.matrix was processed with the RUVseq Bioconductor package102 using the RUVs method to remove low abundance genes, normalize the RNA-seq data, and remove unwanted variation. The RUVseq-processed matrix was then used to identify DE transcripts using both the EdgeR and DESeq2 Bioconductor packages. The resulting lists of DE genes were sorted and filtered to include only those transcripts with false discovery rate (EdgeR) or adjusted p-value (DESeq2) of <0.05 and log2 fold change of >2 or <−2. A list of upregulated and downregulated transcripts that overlapped from both analysis methods was generated and used to create area-proportional Venn diagrams of overlap using BioVenn (http://www.biovenn.nl/) (Fig. 1c).
The retrieved gene lists were analyzed for overrepresented pathways, biological functions, and upstream regulators using IPA (Ingenuity Systems, www.ingenuity.com). p-Values reported for IPA results are generated by IPA using a right-sided Fisher exact test for over-representation analysis, Benjamini–Hochberg correction for multiple hypothesis testing correction, and a z-score algorithm for upstream regulator analysis, p-values < 0.05 were considered significant. For the systems-level analysis, the C. mydas DE transcripts were annotated to their closest characterized human homolog. Human annotation was used to enable the most comprehensive systems-level analysis, as human genes have been the most extensively annotated and characterized.
To detect ChHV5 transcripts independently of our de novo transcriptome analysis, trimmed reads (see above) were aligned to the ChHV5 partial genome [GenBank accession number: HQ878327.2] using HISAT2 (https://ccb.jhu.edu/software/hisat2/index.shtml)103. Aligned non-repetitive reads were blasted (megablast, https://blast.ncbi.nlm.nih.gov/Blast.cgi) against the NCBI nucleotide collection. These reads were specific to ChHV5 and their location within the ChHV5 genome were visualized using Blast’s alignment graphics function (https://goo.gl/YVQrkc).
Additional software tools and datasets: IPA software was used for the ITR, pathway, and GO analysis. String (www.string-db.org) was used to generate protein–protein interaction networks, and the KEGG pathway enrichment analysis tool in String was also applied to these networks. Area-proportional Venn diagrams were generated using BioVenn (http://www.biovenn.nl/). GeneMANIA (v3.5.0, http://genemania.org/)104 was used to generate multiple association networks. UVI data was obtained from publicly available NOOA datasets (http://www.ftp.cpc.ncep.noaa.gov/long/uv/cities/ and http://www.cpc.ncep.noaa.gov/products/stratosphere/uv_index/uv_annual.shtml). Stranding data was obtained from Foley et al.73, with the raw data points generously provided directly to us by Dr. Allen Foley.
RT-qPCR
RNA was extracted as above. Sixty-six tumor and 14 non-tumor RNA samples from five juvenile green turtles (C. mydas), which had stranded in Northern FL were used for cDNA was synthesis using a QuantiTect Reverse Transcription Kit (Qiagen) including genomic DNA digestion. A LightCycler480 Instrument II (Roche) and LightCycler480 SYBR 1 Master reagents (Roche) were used according to the manufacturer’s protocol. Cycling parameters were as follows: 95 °C 10 min, 45 cycles of 95 °C 10 s, 60 °C 20 s, 72 °C 20 s and a melting curve. Primer sequences are provided in Table 1. Gene expression was normalized to the expression of β-actin31 with Rplo (Table 1) as a second endogenous control. Technical replicates for every sample were also performed. Upon admittance to the hospital, the turtles used in the qPCR study had a tumor score range of 2–3 (Hawaii classification system)105, and a Southwest Atlantic fibropapillomatosis (FPSSWA) tumor score range of mild to severe (fibropapillomatosis index, FPI, range, 0.6 to >185.5) (Southwest Atlantic classification system)106.
Data availability
The RNA-seq data was deposited at NCBI (https://www.ncbi.nlm.nih.gov/) under BioProject ID: PRJNA449022 (http://www.ncbi.nlm.nih.gov/bioproject/449022), while the Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL-EBI/GenBankunder the accession GGMX00000000. The version described in this paperis the first version, GGMX01000000.
References
Smith, G. C. & Coates, C. W. Fibro-epithelial growths of the skin in large marine turtles Chelonia mydas. Zoologica 23, 93–98 (1938).
Lucke, B. Studies on tumors in cold-blooded vertebrates. Annual Report of the Tortugas Laboratory of the Camegie Institute 1937, 92–94 (1938).
Cruz Sr, E. (1985). Saga of the sea turtle. FL, USA: Privately-published.
Whilde, J., Martindale, M. Q. & Duffy, D. J. Precision wildlife medicine: applications of the human-centred precision medicine revolution to species conservation. Global Change Biology 23, 1792–1805 (2017).
Jones, K., Ariel, E., Burgess, G. & Read, M. A review of fibropapillomatosis in green turtles (Chelonia mydas). Veterinary Journal 212, 48–57 (2016).
Hargrove, S., Work, T., Brunson, S., Foley, A. M., & Balazs, G. (2016). Proceedings of the 2015 international summit on fibropapillomatosis: global status, trends, and population impacts. In U.S. Dep. Commer., NOAA Technical Memorandum, NOAA-TM-NMFS-PIFSC-054 85.
Page-Karjian, A. et al. Factors influencing survivorship of rehabilitating green sea turtles (Chelonia mydas) with fibropapillomatosis. Journal of Zoo and Wildlife Medicine 45, 507–519 (2014).
Duarte, A. et al. First histological and virological report of fibropapilloma associated with herpesvirus in Chelonia mydas at Príncipe Island, West Africa. Archives of Virology 157, 1155–1159 (2012).
Foley, A. M., Singel, K., Hardy, R., Bailey, R., & Schaf, S. (2007). Distributions, relative abundances, and mortality factors for sea turtles in Florida from 1980 through 2007 as determined from strandings. Florida Fish and Wildlife Conservation Commission.
Herbst, L. H. Fibropapillomatosis of marine turtles. Annual Review of Fish Diseases 4, 389–425 (1994).
Aguirre, A. A. & Lutz, P. Marine turtles as sentinels of ecosystem health: is fibropapillomatosis an indicator? EcoHealth 1, 275–283 (2004).
Williams, E. H. & Bunkley-Williams, L. Early fibropapillomas in Hawaii and occurrences in all sea turtle species: the panzootic, associated leeches wide-ranging on sea turtles, and species of study leeches should be identified. Journal of Virology 80, 4643–4644 (2006).
Page-Karjian, A. et al. Presence of chelonid fibropapilloma-associated herpesvirus in tumored and non-tumored green turtles, as detected by polymerase chain reaction, in endemic and non-endemic aggregations, Puerto Rico. SpringerPlus 1, 35 (2012).
McAloose, D. & Newton, A. L. Wildlife cancer: a conservation perspective. Nature Reviews Cancer 9, 517–526 (2009).
Gil da Costa, R. M., Peleteiro, M. C., Pires, M. A. & DiMaio, D. An update on canine, feline and bovine papillomaviruses. Transboundary and Emerging Diseases 64, 1371–1379 (2016).
Research AAFC. AACR Cancer Progress Report 2012. Clinical Cancer Research 18, S1–S100 (2012).
Davidson, N. E. et al. AACR Cancer Progress Report 2016. Clinical Cancer Research 22, S1–S137 (2016).
Jones, K. E. et al. Global trends in emerging infectious diseases. Nature 451, 990–993 (2008).
Taylor, L. H., Latham, S. M. & Mark, E. Risk factors for human disease emergence. Philosophical Transactions of the Royal Society of London B: Biological Sciences 356, 983–989 (2001).
Wolfe, N. D., Dunavan, C. P. & Diamond, J. Origins of major human infectious diseases. Nature 447, 279–283 (2007).
Woolhouse, M. E., & Gowtage-Sequeria, S. (2006). Host range and emerging and reemerging pathogens. In Ending the war metaphor: the changing agenda for unraveling the host-microbe relationship-Workshop Summary. Vol. 192.
Cleaveland, S., Laurenson, M. & Taylor, L. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philosophical Transactions of the Royal Society of London B: Biological Sciences 356, 991–999 (2001).
Daszak, P., Cunningham, A. A. & Hyatt, A. D. Emerging infectious diseases of wildlife–threats to biodiversity and human health. Science 287, 443–449 (2000).
Daszak, P., Cunningham, A. A. & Hyatt, A. D. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Tropica 78, 103–116 (2001).
Lamb, J. B. & Willis, B. L. Using coral disease prevalence to assess the effects of concentrating tourism activities on offshore reefs in a tropical marine park. Conservation Biology 25, 1044–1052 (2011).
Williams, E. H. Jr et al. An epizootic of cutaneous fibropapillomas in green turtles Chelonia mydas of the Caribbean: part of a panzootic? Journal of Aquatic Animal Health 6, 70–78 (1994).
Herbst, L. et al. Experimental transmission of green turtle fibropapillomatosis using cell-free tumor extracts. Diseases of Aquatic Organisms 22, 1–12 (1995).
Work, T. M. et al. In vitro biology of fibropapilloma-associated turtle herpesvirus and host cells in Hawaiian green turtles (Chelonia mydas). Journal of General Virology 90, 1943–1950 (2009).
Lu, Y. et al. Establishment and characterization of 13 cell lines from a green turtle (Chelonia mydas) with fibropapillomas. In Vitro Cellular & Developmental Biology – Animal 35, 389–393 (1999).
Alfaro-Núñez, A. et al. Further evidence of chelonid herpesvirus 5 (ChHV5) latency: high levels of ChHV5 DNA detected in clinically healthy marine turtles. PeerJ 4, e2274 (2016).
Page-Karjian, A. et al. Quantifying chelonid herpesvirus 5 in symptomatic and asymptomatic rehabilitating green sea turtles. Endangered Species Research 28, 135–146 (2015).
Morrison, C. L. et al. Genomic evolution, recombination, and inter-strain diversity of chelonid alphaherpesvirus 5 from Florida and Hawaii green sea turtles with fibropapillomatosis. PeerJ 6, e4386 (2018).
Ackermann, M. et al. The genome of chelonid herpesvirus 5 harbors atypical genes. PLoS One 7, e46623 (2012).
Chaves, A. et al. Examining the role of transmission of chelonid alphaherpesvirus 5. EcoHealth 14, 530–541 (2017).
Work, T. M., Dagenais, J., Weatherby, T. M., Balazs, G. H. & Ackermann, M. In-vitro replication of chelonid herpesvirus 5 in organotypic skin cultures from Hawaiian green turtles (Chelonia mydas). Journal of Virology 91, e00404–e00417 (2017).
Alfaro-Núñez, A. et al. Global distribution of chelonid fibropapilloma-associated herpesvirus among clinically healthy sea turtles. BMC Evolutionary Biology 14, 1–12 (2014).
Page-Karjian, A. et al. Potential non-cutaneous sites of chelonid herpesvirus 5 persistence and shedding in green sea turtles (Chelonia mydas). Journal of Aquatic Animal Health 29, 136–142 (2017).
Patrício, A. R. et al. Global phylogeography and evolution of chelonid fibropapilloma-associated herpesvirus. Journal of General Virology 93, 1035–1045 (2012).
Herbst, L., Ene, A., Su, M., Desalle, R. & Lenz, J. Tumor outbreaks in marine turtles are not due to recent herpesvirus mutations. Current Biology 14, R697–R699 (2004).
Greenblatt, R. J. et al. Genomic variation of the fibropapilloma-associated marine turtle herpesvirus across seven geographic areas and three host species. Journal of Virology 79, 1125–1132 (2005).
Foley, A. M., Schroeder, B. A., Redlow, A. E., Fick-Child, K. J. & Teas, W. G. Fibropapillomatosis in stranded green turtles (Chelonia mydas) from the eastern United States (1980–98): trends and associations with environmental factors. Journal of Wildlife Diseases 41, 29–41 (2005).
Perrault, J. R. et al. Potential effects of brevetoxins and toxic elements on various health variables in Kemp’s ridley (Lepidochelys kempii) and green (Chelonia mydas) sea turtles after a red tide bloom event. Science of the Total Environment 605, 967–979 (2017).
Van Houtan, K. S., Hargrove, S. K. & Balazs, G. H. Land use, macroalgae, and a tumor-forming disease in marine turtles. PLoS One 5, e12900 (2010).
dos Santos, R. G. et al. Relationship between fibropapillomatosis and environmental quality: a case study with Chelonia mydas off Brazil. Diseases of Aquatic Organisms 89, 87–95 (2010).
Schulz, T. F. & Cesarman, E. Kaposi sarcoma-associated herpesvirus: mechanisms of oncogenesis. Current Opinion in Virology 14, 116–128 (2015).
Niller, H. H., Wolf, H. & Minarovits, J. Viral hit and run-oncogenesis: genetic and epigenetic scenarios. Cancer Letters 305, 200–217 (2011).
Duffy, D. J. Problems, challenges and promises: perspectives on precision medicine. Briefings in Bioinformatics 17, 494–504 (2016).
Jameson, J. L. & Longo, D. L. Precision medicine — personalized, problematic, and promising. New England Journal of Medicine 372, 2229–2234 (2015).
Collins, F. S. & Varmus, H. A new initiative on precision medicine. New England Journal of Medicine 372, 793–795 (2015).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols 8, 1494–1512 (2013).
Das, D. K. et al. miR-1207-3p regulates the androgen receptor in prostate cancer via FNDC1/fibronectin. Experimental Cell Research 348, 190–200 (2016).
Garber, A. C., Hu, J. & Renne, R. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. Journal of Biological Chemistry 277, 27401–27411 (2002).
Boratyn, G. M. et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Research 41, W29–W33 (2013).
Szklarczyk, D. et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Research 45, D362–D368 (2017).
Biankin, A. V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012).
Pardal, R., Clarke, M. F. & Morrison, S. J. Applying the principles of stem-cell biology to cancer. Nature Reviews Cancer 3, 895–902 (2003).
Wahl, G. M. & Spike, B. T. Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. NPJ Breast Cancer 3, 14 (2017).
Barillari, G. et al. The Tat protein of human immunodeficiency virus type-1 promotes vascular cell growth and locomotion by engaging the α5β1 and αvβ3 integrins and by mobilizing sequestered basic fibroblast growth factor. Blood 94, 663–672 (1999).
Krämer, A., Green, J., Pollard, J. & Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 (2014).
Duffy, D. J., Krstic, A., Schwarzl, T., Higgins, D. G. & Kolch, W. GSK3 inhibitors regulate MYCN mRNA levels and reduce neuroblastoma cell viability through multiple mechanisms, including p53 and Wnt signaling. Molecular Cancer Therapeutics 13, 454–467 (2014).
Tortolina, L. et al. Advances in dynamic modeling of colorectal cancer signaling-network regions, a path toward targeted therapies. Oncotarget 6, 5041–5058 (2015).
Duffy, D. J. et al. Wnt signalling is a bi-directional vulnerability of cancer cells. Oncotarget 7, 60310–60331 (2016).
Zhan, T., Rindtorff, N. & Boutros, M. Wnt signaling in cancer. Oncogene 36, 1461–1473 (2017).
Takebe, N. et al. Targeting notch, hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nature Reviews Clinical Oncology 12, 445–464 (2015).
Yang, L., Xie, G., Fan, Q. & Xie, J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene 29, 469–481 (2009).
Wakefield, L. M. & Hill, C. S. Beyond TGF[beta]: roles of other TGF[beta] superfamily members in cancer. Nature Reviews Cancer 13, 328–341 (2013).
Hu, B. et al. Epigenetic activation of WNT5A drives glioblastoma stem cell differentiation and invasive growth. Cell 167, 1281–1295.e18 (2016).
Kumawat, K. & Gosens, R. WNT-5A: signaling and functions in health and disease. Cellular and Molecular Life Sciences 73, 567–587 (2016).
Duffy, D. J. et al. Retinoic acid and TGF-β signalling cooperate to overcome MYCN-induced retinoid resistance. Genome Medicine 9, 15 (2017).
Dyberg, C. et al. Planar cell polarity gene expression correlates with tumor cell viability and prognostic outcome in neuroblastoma. BMC Cancer 16, 1–14 (2016).
Blanc, E., Roux, G. L., Benard, J. & Raguenez, G. Low expression of Wnt-5a gene is associated with high-risk neuroblastoma. Oncogene 24, 1277–1283 (2004).
Corda, G. & Sala, A. Non-canonical WNT/PCP signalling in cancer: Fzd6 takes centre stage. Oncogenesis 6, e364 (2017).
Foley, A. M., et al. (2015). Distributions, relative abundances, and mortality factors of sea turtles in Florida during 1980–2014 as determined from strandings. Jacksonville, FL, USA: Fish and Wildlife Research Institute.
Chang, N.-B., Feng, R., Gao, Z. & Gao, W. Skin cancer incidence is highly associated with ultraviolet-B radiation history. International Journal of Hygiene and Environmental Health 213, 359–368 (2010).
Love, W., Bernhard, J. D. & Bordeaux, J. S. Topical imiquimod or fluorouracil therapy for basal and squamous cell carcinoma: a systematic review. Archives of Dermatology 145, 1431–1438 (2009).
Hallmann, C. A. et al. More than 75 percent decline over 27 years in total flying insect biomass in protected areas. PLoS One 12, e0185809 (2017).
Ceballos, G., Ehrlich, P. R. & Dirzo, R. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proceedings of the National Academy of Sciences 114, E6089–E6096 (2017).
Barnosky, A. D. et al. Has the Earth’s sixth mass extinction already arrived? Nature 471, 51–57 (2011).
Pimm, S. L. et al. The biodiversity of species and their rates of extinction, distribution, and protection. Science 344, 1246752 (2014).
Tilman, D. et al. Future threats to biodiversity and pathways to their prevention. Nature 546, 73–81 (2017).
Heard, M. J. et al. The threat of disease increases as species move toward extinction. Conservation Biology 27, 1378–1388 (2013).
Gore, S. M. et al. Neuronal differentiation in basal cell carcinoma: possible relationship to hedgehog pathway activation? Journal of Pathology 219, 61–68 (2009).
Apalla, Z. et al. Spotlight on vismodegib in the treatment of basal cell carcinoma: an evidence-based review of its place in therapy. Clinical, Cosmetic and Investigational Dermatology 10, 171 (2017).
Nusse, R. & Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Duffy, D. J. et al. Integrative omics reveals MYCN as a global suppressor of cellular signalling and enables network-based therapeutic target discovery in neuroblastoma. Oncotarget 6, 43182–43201 (2015).
Zhou, A. Y. & Johnson, D. B. Combinatorial therapies in melanoma: MAPK inhibitors and beyond. American Journal of Clinical Dermatology 19, 181–193 (2018).
Grimaldi, A. M. et al. MEK inhibitors in the treatment of metastatic melanoma and solid tumors. American Journal of Clinical Dermatology 18, 745–754 (2017).
Zhao, Y. & Adjei, A. A. The clinical development of MEK inhibitors. Nature Reviews Clinical Oncology 11, 385–400 (2014).
Marzagalli, M. et al. Estrogen receptor β in melanoma: from molecular insights to potential clinical utility. Frontiers in Endocrinology 7, 140 (2016).
Colak, S. & ten Dijke, P. Targeting TGF-β signaling in cancer. Trends in Cancer 3, 56–71 (2017).
Brenneisen, P., Sies, H. & Scharffetter‐Kochanek, K. Ultraviolet‐B irradiation and matrix metalloproteinases. Annals of the New York Academy of Sciences 973, 31–43 (2002).
Chandler, H. L., Kusewitt, D. F. & Colitz, C. M. Modulation of matrix metalloproteinases by ultraviolet radiation in the canine cornea. Veterinary Ophthalmology 11, 135–144 (2008).
Dong, K. K. et al. UV‐induced DNA damage initiates release of MMP‐1 in human skin. Experimental Dermatology 17, 1037–1044 (2008).
Krishnapati, L.-S. et al. UV induced foot duplication in regenerating hydra is mediated by metalloproteinases and modulation of the Wnt pathway. International Journal of Developmental Biology 60, 111–117 (2016).
Kripke, M. L., Cox, P. A., Alas, L. G. & Yarosh, D. B. Pyrimidine dimers in DNA initiate systemic immunosuppression in UV-irradiated mice. Proceedings of the National Academy of Sciences 89, 7516–7520 (1992).
Williamson, C. E. et al. Solar ultraviolet radiation in a changing climate. Nature Climate Change 4, 434–441 (2014).
Van Houtan, K. S., Smith, C. M., Dailer, M. L. & Kawachi, M. Eutrophication and the dietary promotion of sea turtle tumors. PeerJ 2, e602 (2014).
Mole, N., Fielding, M. & Lunt, D. The influence of UV disinfection on the formation of disinfection by-products. Disinfection By-products in Drinking Water: Current Issues 245, 54 (1999).
Hunting, E. R. et al. UV radiation and organic matter composition shape bacterial functional diversity in sediments. Frontiers in Microbiology 4, 317 (2013).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology 11, R25 (2010).
Dillies, M.-A. et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Briefings in Bioinformatics 14, 671–683 (2013).
Risso, D., Ngai, J., Speed, T. P. & Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nature Biotechnology 32, 896–902 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nature Methods 12, 357 (2015).
Zuberi, K. et al. GeneMANIA prediction server 2013 update. Nucleic Acids Research 41, W115–W122 (2013).
Work, T. M. & Balazs, G. H. Relating tumor score to hematology in green turtles with fibropapillomatosis in Hawaii. Journal of Wildlife Diseases 35, 804–807 (1999).
Rossi, S. et al. Challenges in evaluating the severity of fibropapillomatosis: a proposal for objective index and score system for green sea turtles (Chelonia mydas) in Brazil. PLoS One 11, e0167632 (2016).
Hulsen, T., de Vlieg, J. & Alkema, W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488 (2008).
Acknowledgments
Funding was generously provided by the Save Our Seas Foundation under project number SOSF 356, The Sea Turtle Conservancy, Florida Sea Turtle Grants Program, project number 17-033R, a Welsh Government Sêr Cymru II and the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 663830-BU115, and a National Science Foundation REU Site Award (DBI-1560356 to Whitney Lab, University of Florida). Warmest thanks to Nancy Condron, Richie Moretti, and the veterinary and rehabilitation staff and volunteers of the Sea Turtle Hospital at Whitney Laboratories and the Turtle Hospital, Marathon for facilitating the study. Thanks also are due to Yanping Zhang and Mei Zhang of UF’s Interdisciplinary Center for Biotechnology Research Core Facilities, Todd Osborne, Elizabeth Fahsbender, and Mya Breitbart for informative discussions, and Florida Fish and Wildlife Conservation Commission’s Meghan Koperski and Allen Foley for valuable assistance with permitting and stranding data access. The funding agencies had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
D.J.D. and M.Q.M. designed and supervised the project; D.J.D and C.Y. generated the data; D.J.D., C.S., A.K., R.T., K.Y., J.W., and L.K. performed data and bioinformatics analysis; B.B., L.K., R.T., D.R., C.E., and B.Z. provided veterinary care, including tumor removal surgeries and diagnostics, and led and conducted the 5-FU treatments. D.J.D. wrote the manuscript; M.Q.M., C.S., and J.W. performed critical reading of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Duffy, D.J., Schnitzler, C., Karpinski, L. et al. Sea turtle fibropapilloma tumors share genomic drivers and therapeutic vulnerabilities with human cancers. Commun Biol 1, 63 (2018). https://doi.org/10.1038/s42003-018-0059-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-018-0059-x
This article is cited by
-
Preparation of recombinant glycoprotein B (gB) of Chelonid herpesvirus 5 (ChHV5) for antibody production and its application for infection detection in sea turtles
Scientific Reports (2022)
-
A subpopulation of green turtle suprabasal epidermal cells are Langerin+ and migrate under in vitro stimulation of the chemokine CCL21
Veterinary Research Communications (2022)
-
Morphologic and physiologic characteristics of green sea turtle (Chelonia mydas) hatchlings in southeastern Florida, USA
Journal of Comparative Physiology B (2022)
-
Environmental DNA monitoring of oncogenic viral shedding and genomic profiling of sea turtle fibropapillomatosis reveals unusual viral dynamics
Communications Biology (2021)
-
Molecular characterization of a marine turtle tumor epizootic, profiling external, internal and postsurgical regrowth tumors
Communications Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.