Abstract
Patients with myelodysplastic syndromes (MDSs) display severe anemia but the mechanisms underlying this phenotype are incompletely understood. Right open-reading-frame kinase 2 (RIOK2) encodes a protein kinase located at 5q15, a region frequently lost in patients with MDS del(5q). Here we show that hematopoietic cell-specific haploinsufficient deletion of Riok2 (Riok2f/+Vav1cre) led to reduced erythroid precursor frequency leading to anemia. Proteomic analysis of Riok2f/+Vav1cre erythroid precursors suggested immune system activation, and transcriptomic analysis revealed an increase in p53-dependent interleukin (IL)-22 in Riok2f/+Vav1cre CD4+ T cells (TH22). Further, we discovered that the IL-22 receptor, IL-22RA1, was unexpectedly present on erythroid precursors. Blockade of IL-22 signaling alleviated anemia not only in Riok2f/+Vav1cre mice but also in wild-type mice. Serum concentrations of IL-22 were increased in the subset of patients with del(5q) MDS as well as patients with anemia secondary to chronic kidney disease. This work reveals a possible therapeutic opportunity for reversing many stress-induced anemias by targeting IL-22 signaling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The original mass spectra may be downloaded from MassIVE (http://massive.ucsd.edu), MSV000085287. The data are directly accessible via ftp://massive.ucsd.edu/MSV000085287/. Raw RNA-seq data are accessible via Gene Expression Omnibus under accession code GSE165467. Source data for all applicable figures (main and extended) are provided with the paper. The remaining data supporting the findings of this study are available from the corresponding authors upon reasonable request. Materials will be provided with material transfer agreements as appropriate. Source data are provided with this paper.
Change history
14 September 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41590-021-01031-y
References
Giagounidis, A. A., Germing, U. & Aul, C. Biological and prognostic significance of chromosome 5q deletions in myeloid malignancies. Clin. Cancer Res. 12, 5–10 (2006).
Haase, D. et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood 110, 4385–4395 (2007).
Hofmann, W. K., Lubbert, M., Hoelzer, D. & Phillip Koeffler, H. Myelodysplastic syndromes. Hematol. J. 5, 1–8 (2004).
Sole, F. et al. Incidence, characterization and prognostic significance of chromosomal abnormalities in 640 patients with primary myelodysplastic syndromes. Grupo Cooperativo Espanol de Citogenetica Hematologica. Br. J. Haematol. 108, 346–356 (2000).
Dutt, S. et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117, 2567–2576 (2011).
Ebert, B. L. et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451, 335–339 (2008).
Kumar, M. S. et al. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q- syndrome. Blood 118, 4666–4673 (2011).
Ribezzo, F. et al. Rps14, Csnk1a1 and miRNA145/miRNA146a deficiency cooperate in the clinical phenotype and activation of the innate immune system in the 5q- syndrome. Leukemia 33, 1759–1772 (2019).
Schneider, R. K. et al. Role of casein kinase 1A1 in the biology and targeted therapy of del(5q) MDS. Cancer Cell 26, 509–520 (2014).
Schneider, R. K. et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat. Med. 22, 288–297 (2016).
Zemp, I. et al. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185, 1167–1180 (2009).
Muto, T. et al. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat. Immunol. 21, 535–545 (2020).
Smith, M. A. et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 21, 640–650 (2019).
Starczynowski, D. T. & Karsan, A. Innate immune signaling in the myelodysplastic syndromes. Hematol. Oncol. Clin. North Am. 24, 343–359 (2010).
Yang, L., Qian, Y., Eksioglu, E., Epling-Burnette, P. K. & Wei, S. The inflammatory microenvironment in MDS. Cell. Mol. Life Sci. 72, 1959–1966 (2015).
Allampallam, K. et al. Measurement of mRNA expression for a variety of cytokines and its receptors in bone marrows of patients with myelodysplastic syndromes. Anticancer Res. 19, 5323–5328 (1999).
Schipperus, M. R. et al. Interleukin-6 and interleukin-1 enhancement of GM-CSF-dependent proliferation of haematopoietic progenitor cells in myelodysplastic syndromes. Br. J. Haematol. 77, 515–522 (1991).
Shao, L. L. et al. TH22 cells as well as TH17 cells expand differentially in patients with early-stage and late-stage myelodysplastic syndrome. PLoS ONE 7, e51339 (2012).
Verhoef, G. E. et al. Measurement of serum cytokine levels in patients with myelodysplastic syndromes. Leukemia 6, 1268–1272 (1992).
Starczynowski, D. T. & Karsan, A. Deregulation of innate immune signaling in myelodysplastic syndromes is associated with deletion of chromosome arm 5q. Cell Cycle 9, 855–856 (2010).
Means, R. T. Jr. Pathogenesis of the anemia of chronic disease: a cytokine-mediated anemia. Stem Cells 13, 32–37 (1995).
Song, M. et al. IRE1α-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 562, 423–428 (2018).
Ferreira-Cerca, S. et al. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1316–1323 (2012).
Royer-Pokora, B. et al. Delineation by molecular cytogenetics of 5q deletion breakpoints in myelodyplastic syndromes and acute myeloid leukemia. Cancer Genet. Cytogenet. 167, 66–69 (2006).
Tang, G. et al. Isolated del(5q) in patients following therapies for various malignancies may not all be clinically significant. Am. J. Clin. Pathol. 144, 78–86 (2015).
Boultwood, J. et al. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood 99, 4638–4641 (2002).
Lai, F. et al. Transcript map and comparative analysis of the 1.5-Mb commonly deleted segment of human 5q31 in malignant myeloid diseases with a del(5q). Genomics 71, 235–245 (2001).
Seita, J. et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS ONE 7, e40321 (2012).
Khajuria, R. K. et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 173, 90–103 (2018).
Myers, S. A. et al. Streamlined protocol for deep proteomic profiling of FAC-sorted cells and its application to freshly isolated murine immune cells. Mol. Cell. Proteom. 18, 995–1009 (2019).
Su, L. K. et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256, 668–670 (1992).
Gronke, K. et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 566, 249–253 (2019).
Kotenko, S. V. et al. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rβ) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes. J. Biol. Chem. 276, 2725–2732 (2001).
KDOQI & National Kidney Foundation. KDOQI clinical practice guidelines and clinical practice recommendations for anemia in chronic kidney disease. Am. J. Kidney Dis. 47, S11–S145 (2006).
Maciejewski, J. P. et al. A pilot study of the recombinant soluble human tumour necrosis factor receptor (p75)-Fc fusion protein in patients with myelodysplastic syndrome. Br. J. Haematol. 117, 119–126 (2002).
Fenaux, P. et al. Luspatercept in patients with lower-risk myelodysplastic syndromes. N. Engl. J. Med. 382, 140–151 (2020).
Suragani, R. N. et al. Transforming growth factor-β superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat. Med. 20, 408–414 (2014).
Mattapallil, M. J. et al. Interleukin 22 ameliorates neuropathology and protects from central nervous system autoimmunity. J. Autoimmun. 102, 65–76 (2019).
Smith, C. L. et al. IL-22 regulates iron availability in vivo through the induction of hepcidin. J. Immunol. 191, 1845–1855 (2013).
Sakamoto, K. et al. IL-22 controls iron-dependent nutritional immunity against systemic bacterial infections. Sci. Immunol. 2, eaai8371 (2017).
Pietras, E. M. et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18, 607–618 (2016).
Yamashita, M. & Passegue, E. TNF-α coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell 25, 357–372 e357 (2019).
Zhang, T. Y. et al. IL-6 blockade reverses bone marrow failure induced by human acute myeloid leukemia. Sci. Transl. Med. 12, eaax5104 (2020).
Cai, T. et al. Increased expression of IL-22 is associated with disease activity in Behcet’s disease. PLoS ONE 8, e59009 (2013).
Yamamoto-Furusho, J. K. et al. Colonic epithelial upregulation of interleukin 22 (IL-22) in patients with ulcerative colitis. Inflamm. Bowel Dis. 16, 1823 (2010).
Lee, S. J. et al. Certain autoimmune manifestations are associated with distinctive karyotypes and outcomes in patients with myelodysplastic syndrome: a retrospective cohort study. Medicine (Baltimore) 95, e3091 (2016).
Fozza, C., La Nasa, G. & Caocci, G. The Yin and Yang of myelodysplastic syndromes and autoimmunity: The paradox of autoimmune disorders responding to therapies specific for MDS. Crit. Rev. Oncol. Hematol. 142, 51–57 (2019).
Wolach, O. & Stone, R. Autoimmunity and Inflammation in myelodysplastic syndromes. Acta Haematol. 136, 108–117 (2016).
Schnatter, A. R., Glass, D. C., Tang, G., Irons, R. D. & Rushton, L. Myelodysplastic syndrome and benzene exposure among petroleum workers: an international pooled analysis. J. Natl Cancer Inst. 104, 1724–1737 (2012).
Boitano, A. E. et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329, 1345–1348 (2010).
Skarnes, W. C. et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011).
Farley, F. W., Soriano, P., Steffen, L. S. & Dymecki, S. M. Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110 (2000).
Heinrich, A. C., Pelanda, R. & Klingmüller, U. A mouse model for visualization and conditional mutations in the erythroid lineage. Blood 104, 659–666 (2004).
Savage, A. K., Liang, H. E. & Locksley, R. M. The development of steady-state activation hubs between adult LTi ILC3s and primed macrophages in small intestine. J. Immunol. 199, 1912–1922 (2017).
Pronk, C. J. et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1, 428–442 (2007).
Shuga, J., Zhang, J., Samson, L. D., Lodish, H. F. & Griffith, L. G. In vitro erythropoiesis from bone marrow-derived progenitors provides a physiological assay for toxic and mutagenic compounds. Proc. Natl Acad. Sci. USA 104, 8737–8742 (2007).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Shalek, A. K. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 323 (2011).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Dudakov, J. A., Hanash, A. M. & van den Brink, M. R. Interleukin-22: immunobiology and pathology. Annu. Rev. Immunol. 33, 747–785 (2015).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Nikolsky, Y., Ekins, S., Nikolskaya, T. & Bugrim, A. A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol. Lett. 158, 20–29 (2005).
Pellagatti, A. et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia 24, 756–764 (2010).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–D995 (2013).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Acknowledgements
We thank R. Caspi (National Eye Institute, US NIH) and Genentech for providing Il22 knockout mice; B. L. Ebert (DFCI) for providing Rps14-floxed mice; R. Locksley (University of California San Francisco) for providing Catch-22 mice; and U. Klingmüller (Deutsches Krebsforschungszentrum) for providing Epor-cre mice. We also thank the Animal Resources Facility, the Ted and Eileen Pasquarello Tissue Bank in Hematologic Malignancies and flow cytometry core facility at DFCI for their valuable technical assistance. We thank E. Smith for assistance with creating the graphical illustration. We thank P. Májek (Ústav hematologie a krevní transfuse) for providing plasma proteomic profiling data of healthy controls and patients with MDSs. This work was supported by a Discovery Research Grant from the Edward P. Evans Foundation (to L.H.G.) and institutional funding from DFCI (to L.H.G). D.P.S. is supported by the Edward P. Evans Foundation and by NIH Leukemia SPORE 1P50CA206963 and 2P01CA066996. This work was supported in part by grants from the National Cancer Institute (NCI) Clinical Proteomic Tumor Analysis Consortium grants NIH/NCI U24-CA210986 and NIH/NCI U01 CA214125 (to S.A.C.).
Author information
Authors and Affiliations
Contributions
M.R. and L.H.G. conceived the study, designed experiments, analyzed the data and wrote the manuscript; M.R. performed the experiments; S.G. assisted with in vitro experiments; S.A.M. performed proteomic profiling and data analysis; M.S.C. performed RNA-seq; M.S. processed RNA-seq data; S.A.C. analyzed proteomic profiling data; S.S.W. and J.V.B. collected CKD patient samples and related clinical information and analyzed the data; J.R., R.M.S. and D.P.S. collected MDS patient samples and related clinical information and analyzed the data; and A.R. supervised RNA-seq experiments and analyzed data.
Corresponding authors
Ethics declarations
Competing interests
An invention disclosure has been filed based on the data generated in this study. From 4 August 2020, Meromit Singer has been an employee of Guardant Health. S.S.W. has served on the steering committee of a GSK trial on an oral hypoxia-inducible factor prolyl hydroxylase inhibitor, as a potential treatment for anemia associated with CKD; S.S.W. has also received consulting fees from Public Health Advocacy Institute, CVS, Roth Capital Partners, Kantum Pharma, Mallinckrodt, Wolters Kluewer, GE Health Care, Allena Pharmaceuticals, Mass Medical International, JNJ, Venbio, Strataka, Takeda, Cerus and Pfizer. D.P.S. has served on independent data safety monitoring committees for clinical trials supported by Takeda, Astex, Janssen and Onconova; has consulted for Celgene and Daiichi Sankyo; and has received research support (to the institution) for clinical trials sponsored by Aprea, H3 Biosciences, Syros and Astra Zeneca. J.R. reports research funding from Amgen, Equillium and Kite Pharma; and consulting income from Aleta Biotherapeutics, Avrobio, Celgene, Falcon Therapeutics, LifeVault Bio, Rheos Medicines, Talaris Therapeutics and TScan Therapeutics. R.M.S. has served on independent data safety monitoring committees for trials supported by Celgene, Takeda and Argenix; has consulted for AbbVie, Actinium, Agios, Amgen, Arog, Astellas, Astra Zeneca, Biolinerx, Celgene, Daiichi Sankyo, Fujifilm, Janssen, Juno, Macrogenics, Novartis, Ono, Orsenix, Pfizer, Roche, Stemline, Sumitomo, Takeda and Trovagene; and has received research support (to the institution) for clinical trials sponsored by AbbVie, Agios, Arog and Novartis. S.A.C. is a member of the scientific advisory boards of Kymera, PTM BioLabs and Seer and is a scientific advisor to Pfizer and Biogen. A.R. is a Scientific Advisory Board member of Thermo Fisher Scientific, Neogene Therapeutics, Asimov and Syros Pharmaceuticals. A.R. is a cofounder of and equity holder in Celsius Therapeutics and an equity holder in Immunitas. From 1 August 2020, A.R. has been an employee of Genentech. L.H.G. is a former Director of Bristol-Myers Squibb and the Waters Corporation and is currently on the board of directors of and holds equity in GlaxoSmithKline Pharmaceuticals and Analog Devices. She also serves on the scientific advisory boards of Repare Therapeutics, Abpro Therapeutics and Kaleido Therapeutics. S.A.C. is a member of the scientific advisory boards of Kymera, PTM BioLabs, and Seer and is a scientific advisor to Pfizer and Biogen. All other authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks Michaëla Fontenay, Rebekka Schneider and the other, anonymous reviewer(s) for their contribution to the peer review of this work. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Localization and expression of Riok2.
a, Location of RIOK2 gene on human chromosome 5. b, Schematic representation of the Riok2tm1a(KOMP)Wtsi allele and generation of Riok2 floxed mice. c, Agarose gel showing genotyping of Riok2 floxed mice. Riok2Δ indicates deletion of Riok2. No band expected in the Riok2wt lane. d, Expression of Riok2 mRNA in mouse BM cells. Modified from Gene Expression Commons. Numbers next to bars indicate expression level. e, Riok2 mRNA expression by qRT-PCR in BM cells from Riok2 haploinsufficient mice and Vav1cre controls. n = 5 mice/group. f, Frequency of the genotypes indicated on the X-axis among 4 litters from 4 different breeding crosses of the genotypes mentioned. g, In vivo protein synthesis rates in the indicated cell types from Riok2 haploinsufficient mice (n = 2) and Vav1cre controls (n = 8). Unpaired two-tailed t-test (e), multiple unpaired two-tailed t-tests with Holm-Sidak method (g) used to calculate statistical significance. Data are shown as mean ± s.e.m (e,f) or mean ± s.d. g, and are representative of two (e, g) or four (c, f) independent experiments. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
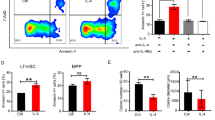
Extended Data Fig. 2 Riok2 haploinsufficient mice display anemia and myeloproliferation.
a, Gating strategy used for the identification of erythroid progenitor/ precursor cells in the BM. b, Number of erythroid progenitor populations among viable BM cells in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls. n = 5/group. c, Cell cycle analysis of erythroid progenitor/ precursor cells from Riok2 haploinsufficient mice in comparison to Vav1cre controls. n = 5 mice/group. d, Cdkn1a mRNA expression by qRT-PCR in erythroid progenitors from Riok2 haploinsufficient mice and Vav1cre controls. n = 3 mice/group. e, Kaplan-Meier survival curve for Riok2 haploinsufficient mice and Vav1cre controls subjected to lethal dose of PhZ. f, Number of RIII and RIV erythroid precursor populations among viable BM cells in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls day 6 after PhZ treatment. n = 4/group. g, PB RBC numbers, Hb, and HCT in mice transplanted with either Riok2 haploinsufficient mice or Vav1cre BM cells. n = 5 mice/group. h, PB RBC numbers, Hb, and HCT in mice with tamoxifen-inducible deletion of Riok2. Tamoxifen administered on days 3 – 7. n = 8 and 7 for Riok2+/+Ert2cre and Riok2f/+Ert2cre mice, respectively. i, Representative flow cytometry plots showing frequency of monocytes (CD11b+Ly6G−Ly6Chi) and neutrophils (CD11b+Ly6G+) in the PB of Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. j, Representative flow cytometry plots showing Ki-67+ GMPs in the BM of Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. k, Number of CFU-GM colonies in MethoCult from Lin−Sca-1+c-kit+ BM cells from Riok2f/+Vav1cre mice (n = 5) and Riok2+/+Vav1cre controls (n = 4) after a 7-day culture period. n = 4-5/group. Multiple unpaired two-tailed t-tests with Holm-Sidak method (b, c), unpaired two-tailed t-test (d, f, g, k), log-rank test (e), and 2-way ANOVA with Sidak’s correction for multiple comparisons (h) used to calculate statistical significance. Data are shown as mean ± s.e.m and are representative of two (b to k) independent experiments. * p<0.05, ** p<0.01, *** p<0.001.
Extended Data Fig. 3 Riok2 haploinsufficiency alters early hematopoietic progenitors in an age-dependent fashion.
a, Frequency and number of indicated cell types in the bone marrow of Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. n = 4/group. LT-HSC=long term hematopoietic stem cells, ST-HSC=short term hematopoietic stem cells, MPP=multipotent progenitors, CLP=common lymphoid progenitors. b, % CD45.2 (donor) chimerism in PB from competitive BM transplant with CD45.1 recipient cells. Time point ‘-1’ reflects first bleeding 4 weeks after transplantation and one day before tamoxifen induced deletion of Riok2. Donor (CD45.2) chimerism of the HSC compartment in the BM of competitive transplantation experiments. n = 5/group. c, Frequency of donor (CD45.2+) early hematopoietic progenitors 24 weeks after tamoxifen treatment in a competitive transplantation assay as described in (b). n = 5 and 4 for Riok2+/+Ert2cre and Riok2f/+Ert2cre mice, respectively. Unpaired two-tailed t-test (a, c) and 2-way ANOVA with Sidak’s multiple comparison test (b) used to calculate statistical significance. Data are shown as mean ± s.e.m and are representative of two (a-c) independent experiments. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Extended Data Fig. 4 Riok2 haploinsufficient erythroid precursors express increased S100 proteins.
(a) Expression of ribosomal proteins quantified by proteomics in Riok2f/+Vav1cre and Riok2+/+Vav1cre erythroid precursors. S100A8 (b) and S100A9 (c) expression assessed by flow cytometry in BM erythroid precursors from Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. n = 4mice/group. d,e, S100a8 and S100a9 mRNA expression in erythroid precursors isolated from Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. n = 4 mice/group. f, p53 expression assessed by flow cytometry in BM erythroid precursors from Riok2f/+Vav1cre and Riok2+/+Vav1cre mice. g, Graphical representation of data shown in (e). n = 5 mice/group. Data are shown as mean ± s.e.m and are representative of two (b to g) independent experiments. Unpaired two-tailed t-test (b to g) used to calculate statistical significance. ** p<0.01, *** p<0.001, **** p<0.0001.
Extended Data Fig. 5 Expression of lineage-associated T cell cytokines is comparable between Riok2 haploinsufficient and sufficient T cells.
a-g, Concentration of IL-2 (a), IFN-γ (b), IL-4 (c), IL-5 (d), IL-13 (e), IL-17A (f) and frequency of Foxp3+ cells (g) from in vitro polarized T cells of the indicated genotypes. n = 3 mice/group. h-i) Number of IL-22+ NKT cells (H) and ILCs (I) in the spleens of Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls (n = 4/group). j, Frequency of IL_23p19+ DCs in Riok2f/+Vav1cre mice and Riok2+/+Vav1cre controls. n = 4 mice/group. k, PB RBC numbers, Hb, and HCT in Riok2f/fCd4cre mice (n = 3) in comparison to Riok2+/+Cd4cre controls (n = 5). l, Secreted IL-22 from in vitro polarized TH22 cells from Rps14 haploinsufficient mice and Vav1cre controls. n = 4 mice/group. m, Secreted IL-22 from in vitro polarized TH22 cells from ApcMin mice and littermate controls. n = 4 mice/group. n, Viable cells (expressed as percentage of total cells in culture) for the indicated treatments assessed by flow cytometry. n = 5 mice/group. Data are shown as mean ± s.e.m and are representative of two (a to n) independent experiments. Unpaired two-tailed t-test (a to n) used to calculate statistical significance. * p < 0.05, ** p < 0.01.
Extended Data Fig. 6 Neutralization of IL-22 signaling increases number of erythroid precursors.
a-d, Number of RI-RIV erythroid populations among viable BM cells in the indicated strains undergoing PhZ-induced stress erythropoiesis. For (a), n = 5,5,4, and 4 for Riok2+/+Il22+/+Vav1cre, Riok2+/+Il22+/-Vav1cre, Riok2f/+Il22+/+Vav1cre, Riok2f/+Il22+/-Vav1cre, respectively. For (d), n = 5/group. Data are shown as mean ± s.e.m and are representative of three (a, c) or two (b, d) independent experiments. 1-way ANOVA with Tukey’s correction (a, c) or unpaired two-tailed t-test (b, d) used to calculate statistical significance. * p < 0.05, ** p < 0.01.
Extended Data Fig. 7 IL-22 neutralization alleviates anemia in wt mice undergoing PhZ-induced stress erythropoiesis.
a, PB RBC numbers, Hb, and HCT in naïve wt C57BL/6 J mice treated with isotype control (Rat IgG2aκ, 50 mg/mouse) or anti-IL-22 antibody (50 mg/mouse). n = 5 mice/group. b, PB RBC numbers, Hb, and HCT in wt C57BL/6 J mice undergoing PhZ-induced stress erythropoiesis treated with isotype control or anti-IL-22 antibody. n = 5 mice/group. c, Percentage of RI-RIV erythroid precursors in the BM of mice treated as in (b). n = 4 and 5 for Il22ra1+/+Eporcre and Il22ra1f/fEporcre mice, respectively. Data are shown as mean ± s.e.m and are representative of three (a, b) or two (c) independent experiments. Unpaired two-tailed t-test (a to c) used to calculate statistical significance. * p<0.05, *** p<0.001.
Extended Data Fig. 8 Erythroid precursors express IL-22RA1.
a, Gating strategy employed for assessing IL-22RA1 expression on erythroid precursors. b, Gating strategy to show that majority of IL-22RA1+ cells in the mouse BM are erythroid precursors. c, IL-22RA1 expression on erythroid precursors assessed using flow cytometry and a second antibody targeting a different epitope of IL-22RA1. d, Il22ra1 mRNA expression in the indicated cell types assessed by qRT-PCR. T cells and liver represent negative and positive controls, respectively. n = 4 mice/group. Data are shown as mean ± s.e.m (d) and are representative of three (a to c) or two (d) independent experiments.
Extended Data Fig. 9 Increased IL-22 and its signature genes in del(5q) MDS subjects.
a, Representative flow cytometry plots showing frequency of CD4+IL-22+ cells among total PBMCs in the peripheral blood of MDS patients and healthy subjects. Pre-gated on viable CD3ε+CD4+ cells. Cumulative data shown in Fig. 4e. b, Expression of indicated IL-22 signature genes in CD34+ cells from healthy controls and del(5q) and non−del(5q) MDS patients. n = 17, 47, and 136 for healthy, del(5q) MDS, and non-del(5q) MDS, respectively. Kruskal-Wallis test with Dunn’s correction for multiple comparisons (b) used to calculate statistical significance * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. Solid lines represent median and dashed lines represent quartiles (b).
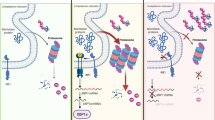
Extended Data Fig. 10 Riok2 haploinsufficiency recapitulates del(5q) MDS transcriptional changes.
a,b, GSEA enrichment plots comparing proteins up-regulated (a) and down-regulated (b) upon Riok2 haploinsufficiency to the transcriptional changes seen in del(5q) MDS. c, Schematic of mechanism underlying Riok2 haploinsufficiency-induced, IL-22 –induced anemia.
Supplementary information
Supplementary Information
Supplementary Tables 1–4.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
About this article
Cite this article
Raundhal, M., Ghosh, S., Myers, S.A. et al. Blockade of IL-22 signaling reverses erythroid dysfunction in stress-induced anemias. Nat Immunol 22, 520–529 (2021). https://doi.org/10.1038/s41590-021-00895-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-021-00895-4
This article is cited by
-
Made to order: emergency myelopoiesis and demand-adapted innate immune cell production
Nature Reviews Immunology (2024)
-
Identification of RIOK2 as a master regulator of human blood cell development
Nature Immunology (2022)
-
There won’T be blood
Nature Immunology (2021)
-
IL-22 blockade alleviates anaemia
Nature Reviews Drug Discovery (2021)