Abstract
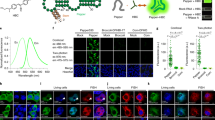
Imaging RNA in living cells is a challenging problem in cell biology. One strategy for genetically encoding fluorescent RNAs is to express them as fusions with Spinach, an 'RNA mimic of GFP'. We found that Spinach was dimmer than expected when used to tag constructs in living cells owing to a combination of thermal instability and a propensity for misfolding. Using systematic mutagenesis, we generated Spinach2 that overcomes these issues and can be used to image diverse RNAs. Using Spinach2, we detailed the dynamics of the CGG trinucleotide repeat–containing 'toxic RNA' associated with Fragile X–associated tremor/ataxia syndrome, and show that these RNAs form nuclear foci with unexpected morphological plasticity that is regulated by the cell cycle and by small molecules. Together, these data demonstrate that Spinach2 exhibits improved versatility for fluorescently labeling RNAs in living cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Medioni, C., Mowry, K. & Besse, F. Principles and roles of mRNA localization in animal development. Development 139, 3263–3276 (2012).
Jansen, R.P. & Niessing, D. Assembly of mRNA-protein complexes for directional mRNA transport in eukaryotes–an overview. Curr. Protein Pept. Sci. 13, 284–293 (2012).
Paige, J.S., Wu, K.Y. & Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 333, 642–646 (2011).
Sellier, C. et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 29, 1248–1261 (2010).
Paige, J.S., Nguyen-Duc, T., Song, W. & Jaffrey, S.R. Fluorescence imaging of cellular metabolites with RNA. Science 335, 1194 (2012).
Ponchon, L. & Dardel, F. Recombinant RNA technology: the tRNA scaffold. Nat. Methods 4, 571–576 (2007).
Prasanth, K.V. et al. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol. Biol. Cell 21, 4184–4196 (2010).
Fu, X.D. & Maniatis, T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343, 437–441 (1990).
Wojciechowska, M. & Krzyzosiak, W.J. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 20, 3811–3821 (2011).
Fu, Y.H. et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67, 1047–1058 (1991).
Dombrowski, C. et al. Premutation and intermediate-size FMR1 alleles in 10572 males from the general population: loss of an AGG interruption is a late event in the generation of fragile X syndrome alleles. Hum. Mol. Genet. 11, 371–378 (2002).
Hagerman, R.J. et al. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 57, 127–130 (2001).
Tassone, F., Iwahashi, C. & Hagerman, P.J. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS). RNA Biol. 1, 103–105 (2004).
Iwahashi, C.K. et al. Protein composition of the intranuclear inclusions of FXTAS. Brain 129, 256–271 (2006).
Sellier, C. et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in Fragile X–associated tremor/ataxia syndrome. Cell Rep. 3, 869–880 (2013).
Todd, P.K. et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455 (2013).
Kiliszek, A., Kierzek, R., Krzyzosiak, W.J. & Rypniewski, W. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 39, 7308–7315 (2011).
Stoss, O. et al. The STAR/GSG family protein rSLM-2 regulates the selection of alternative splice sites. J. Biol. Chem. 276, 8665–8673 (2001).
Gossen, M. & Bujard, H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89, 5547–5551 (1992).
Sobczak, K., de Mezer, M., Michlewski, G., Krol, J. & Krzyzosiak, W.J. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 31, 5469–5482 (2003).
Disney, M.D. et al. A small molecule that targets r(CGG)exp and improves defects in fragile X–associated tremor ataxia syndrome. ACS Chem. Biol. 7, 1711–1718 (2012).
Suganuma, M. et al. Tautomycin: an inhibitor of protein phosphatases 1 and 2A but not a tumor promoter on mouse skin and in rat glandular stomach. J. Cancer Res. Clin. Oncol. 121, 621–627 (1995).
Cohen, P., Klumpp, S. & Schelling, D.L. An improved procedure for identifying and quantitating protein phosphatases in mammalian tissues. FEBS Lett. 250, 596–600 (1989).
Genové, G., Glick, B.S. & Barth, A.L. Brighter reporter genes from multimerized fluorescent proteins. Biotechniques 39, 814–818 (2005).
Acknowledgements
We thank K.Y. Wu for her role in developing early improvements in Spinach, G.S. Filonov, W. Song, N. Svensen and J. Paige for useful comments and suggestions, and F. Dardel (Université Paris Descartes) for providing plasmids containing the tRNA scaffold sequence. This work was supported by US National Institutes of Health NINDS NS010249 (S.R.J.), NIGMS F32 GM106683 (R.L.S.), and NIGMS GM079235 (M.D.D.).
Author information
Authors and Affiliations
Contributions
R.L.S., M.D.D. and S.R.J. conceived and designed the experiments, M.D.D. provided compounds and assisted in experiments using 1a and CGG foci, R.L.S. performed experiments and analyzed data, and R.L.S. and S.R.J. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.R.J. and R.L.S. are authors of a patent application (provisional patent USPTO# 61/874,819) related to technology described in this paper.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1 and 2, and Supplementary Note (PDF 1314 kb)
Formation of foci after transient transfection.
COS-7 cells were transfected with a plasmid that expresses (CGG)60-Spinach2. At 2 h post-transfection, cells were changed into imaging medium containing 20 μM DFHBI. Images were acquired every 20 min for 6 h. The cell in the lower left side of the image displays initial nucleoplasmic signal, followed by formation of smaller foci that grow larger and brighter throughout the experiment. (MOV 779 kb)
Disaggregation of foci after treatment with tautomycin.
A COS-7 cell expressing (CGG)60-Spinach2 aggregates was treated with 5 μM tautomycin and imaged every 5 min for 2 h. Foci disaggregate, leading to nucleoplasmic (CGG)60-Spinach2 after ∼1 h. (MOV 232 kb)
Rights and permissions
About this article
Cite this article
Strack, R., Disney, M. & Jaffrey, S. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat–containing RNA. Nat Methods 10, 1219–1224 (2013). https://doi.org/10.1038/nmeth.2701
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2701
This article is cited by
-
Avidity-based bright and photostable light-up aptamers for single-molecule mRNA imaging
Nature Chemical Biology (2023)
-
Fast-exchanging spirocyclic rhodamine probes for aptamer-based super-resolution RNA imaging
Nature Communications (2023)
-
Nucleic acid-based fluorescent sensor systems: a review
Polymer Journal (2022)
-
Spinach-based RNA mimicking GFP in plant cells
Functional & Integrative Genomics (2022)
-
Intracellular mRNA transport and localized translation
Nature Reviews Molecular Cell Biology (2021)