Abstract
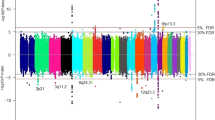
We conducted a two-stage genome-wide association study of renal cell carcinoma (RCC) in 3,772 affected individuals (cases) and 8,505 controls of European background from 11 studies and followed up 6 SNPs in 3 replication studies of 2,198 cases and 4,918 controls. Two loci on the regions of 2p21 and 11q13.3 were associated with RCC susceptibility below genome-wide significance. Two correlated variants (r2 = 0.99 in controls), rs11894252 (P = 1.8 × 10−8) and rs7579899 (P = 2.3 × 10−9), map to EPAS1 on 2p21, which encodes hypoxia-inducible-factor-2 alpha, a transcription factor previously implicated in RCC. The second locus, rs7105934, at 11q13.3, contains no characterized genes (P = 7.8 × 10−14). In addition, we observed a promising association on 12q24.31 for rs4765623, which maps to SCARB1, the scavenger receptor class B, member 1 gene (P = 2.6 × 10−8). Our study reports previously unidentified genomic regions associated with RCC risk that may lead to new etiological insights.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ferlay, J., Bray, F., Pisani, P. & Parkin, D.M. GLOBOCAN 2002: Cancer Incidence, Mortality and Prevalence Worldwide. (IARC CancerBase No. 5. version 2.0, IARCPress, Lyon, France, 2004).
Scélo, G. & Brennan, P. The epidemiology of bladder and kidney cancer. Nat. Clin. Pract. Urol. 4, 205–217 (2007).
Chow, W.H., Dong, L.M. & Devesa, S.S. Epidemiology and risk factors for kidney cancer. Nat. Rev. Urol. 7, 245–257 (2010).
McLaughlin, J.K. et al. A population–based case–control study of renal cell carcinoma. J. Natl. Cancer Inst. 72, 275–284 (1984).
Schlehofer, B. et al. International renal-cell-cancer study. VI. The role of medical and family history. Int. J. Cancer 66, 723–726 (1996).
Gago-Dominguez, M., Yuan, J.M., Castelao, J.E., Ross, R.K. & Yu, M.C. Family history and risk of renal cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 10, 1001–1004 (2001).
Hung, R.J. et al. Family history and the risk of kidney cancer: a multicenter case-control study in Central Europe. Cancer Epidemiol. Biomarkers Prev. 16, 1287–1290 (2007).
Linehan, W.M. et al. Hereditary kidney cancer: unique opportunity for disease-based therapy. Cancer 115, 2252–2261 (2009).
Peto, J. & Houlston, R.S. Genetics and the common cancers. Eur. J. Cancer 37 Suppl 8, S88–S96 (2001).
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Frazer, K.A. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Packer, B.R. et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 34, D617–D621 (2006).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Skol, A.D., Scott, L.J., Abecasis, G.R. & Boehnke, M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38, 209–213 (2006).
Higgins, J.P. et al. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am. J. Pathol. 162, 925–932 (2003).
Xia, G. et al. Regulation of vascular endothelial growth factor transcription by endothelial PAS domain protein 1 (EPAS1) and possible involvement of EPAS1 in the angiogenesis of renal cell carcinoma. Cancer 91, 1429–1436 (2001).
Sowter, H.M., Raval, R.R., Moore, J.W., Ratcliffe, P.J. & Harris, A.L. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res. 63, 6130–6134 (2003).
Kondo, K., Kim, W.Y., Lechpammer, M. & Kaelin, W.G. Jr. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1, E83 (2003).
Zimmer, M., Doucette, D., Siddiqui, N. & Iliopoulos, O. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol. Cancer Res. 2, 89–95 (2004).
Gunaratnam, L. & Bonventre, J.V. HIF in kidney disease and development. J. Am. Soc. Nephrol. 20, 1877–1887 (2009).
Chanock, S. High marks for GWAS. Nat. Genet. 41, 765–766 (2009).
Thomas, G. et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 40, 310–315 (2008).
Eeles, R.A. et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat. Genet. 40, 316–321 (2008).
Turnbull, C. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 42, 504–507 (2010).
Kozarsky, K.F. et al. Overexpression of the HDL receptor SR-BI alters plasma HDL and bile cholesterol levels. Nature 387, 414–417 (1997).
Rigotti, A. et al. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc. Natl. Acad. Sci. USA 94, 12610–12615 (1997).
Ueda, Y. et al. Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B type I transgenic mice. J. Biol. Chem. 274, 7165–7171 (1999).
Yeager, M. et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 39, 645–649 (2007).
Hunter, D.J. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 39, 870–874 (2007).
Landi, M.T. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 85, 679–691 (2009).
Amundadottir, L. et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 41, 986–990 (2009).
Petersen, G.M. et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 42, 224–228 (2010).
Yu, K. et al. Population substructure and control selection in genome-wide association studies. PLoS ONE 3, e2551 (2008).
Falush, D., Stephens, M. & Pritchard, J.K. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7, 574–578 (2007).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
de Bakker, P.I. et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 17, R122–R128 (2008).
Aulchenko, Y.S., Struchalin, M.V. & van Duijn, C.M. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11, 134 (2010).
Acknowledgements
The authors thank all of the participants who took part in this research and the funders and support staff who made this study possible. Funding for the genome-wide genotyping was provided by the French Institut National du Cancer (INCa) for those studies coordinated by IARC/CNG, and by the intramural research program of the National Cancer Institute (NCI), US National Institutes of Health (NIH) for those studies coordinated by the NCI. Additional acknowledgments can be found in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
M.P.P., M.J., J.R.T., G.S., L.E.M., V.G., W.-H.C., J.D.M., N.R., S.J.C. and P. Brennan contributed to the design and execution of the overall study. M.P.P., M.J., J.R.T., G.S., L.E.M., L.A.K., X.W., V.G., K.B.J., J.D.M., N.R., S.J.C. and P. Brennan contributed to the statistical analyses. M.P.P., M.J., S.J.C. and P. Brennan wrote the first draft of the manuscript. D. Zelenika, E.P., L.A.K., X.W., K.B.J., S.H.V., S.L.v.d.M., Y.Y., A.M.M., E.S.B., N.N.C., M.F., D.L., I.G., S.H., H. Blanche, A.H., G.S.T., Z.W., M.Y., K.G.S., S.J.C. and M.L. supervised or conducted the genotyping. The remaining authors conducted the epidemiologic studies and contributed samples to the GWAS and/or replication studies. All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1, 3 and 4, Supplementary Figures 1–3 and Supplementary Note (PDF 677 kb)
Supplementary Table 2
Association results for SNPs imputed on 2p21 (EPAS1), 11q13.3, and 12q24.31 (SCARB1), using data from 1000 Genomes as scaffold (XLS 7053 kb)
Rights and permissions
About this article
Cite this article
Purdue, M., Johansson, M., Zelenika, D. et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet 43, 60–65 (2011). https://doi.org/10.1038/ng.723
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.723
This article is cited by
-
Long-term treatment of chronic kidney disease patients with anemia using hypoxia-inducible factor prolyl hydroxylase inhibitors: potential concerns
Pediatric Nephrology (2024)
-
STAT3 phosphorylation at serine 727 activates specific genetic programs and promotes clear cell renal cell carcinoma (ccRCC) aggressiveness
Scientific Reports (2023)
-
Association between circulating vitamin E and ten common cancers: evidence from large-scale Mendelian randomization analysis and a longitudinal cohort study
BMC Medicine (2022)
-
Potential DNA methylation biomarkers for the detection of clear cell renal cell carcinoma identified by a whole blood-based epigenome-wide association study
Epigenetics Communications (2022)
-
Lipid metabolism reprogramming in renal cell carcinoma
Cancer and Metastasis Reviews (2022)