Abstract
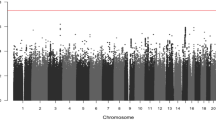
Wilms tumor is the most common renal malignancy of childhood. To identify common variants that confer susceptibility to Wilms tumor, we conducted a genome-wide association study in 757 individuals with Wilms tumor (cases) and 1,879 controls. We evaluated ten SNPs in regions significantly associated at P < 5 × 10−5 in two independent replication series from the UK (769 cases and 2,814 controls) and the United States (719 cases and 1,037 controls). We identified clear significant associations at 2p24 (rs3755132, P = 1.03 × 10−14; rs807624, P = 1.32 × 10−14) and 11q14 (rs790356, P = 4.25 × 10−15). Both regions contain genes that are plausibly related to Wilms tumorigenesis. We also identified candidate association signals at 5q14, 22q12 and Xp22.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Change history
06 June 2012
In the version of this article initially published, the name of one of the authors was incorrectly listed as James Nicholdson. The correct name is James Nicholson. The error has been corrected in the HTML and PDF versions of the article.
10 July 2013
In the version of this article initially published, the following statement was omitted from the Acknowledgments: "We acknowledge use of data from the database of Genotypes and Phenotypes (dbGaP) from the NCBI, US National Library of Medicine. Research support to collect data and develop an application to support High Density SNP Association Analysis of Melanoma (phs000187) was provided by grants 3P50CA093459, 5P50CA097007, 5R01ES011740 and 5R01CA133996 from the US National Institutes of Health. This study also used, in part, data from the National Institute of Neurological Disorders and Stroke (NINDS) dbGaP database from the CIDR:NGRC Parkinson's Disease Study (phs000196)." The error has been corrected in the HTML and PDF versions of the article.
References
Stiller, C.A. & Parkin, D.M. International variations in the incidence of childhood renal tumours. Br. J. Cancer 62, 1026–1030 (1990).
Breslow, N.E., Beckwith, J.B., Perlman, E.J. & Reeve, A.E. Age distributions, birth weights, nephrogenic rests, and heterogeneity in the pathogenesis of Wilms tumor. Pediatr. Blood Cancer 47, 260–267 (2006).
Scott, R.H., Stiller, C.A., Walker, L. & Rahman, N. Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J. Med. Genet. 43, 705–715 (2006).
Scott, R.H. et al. Constitutional 11p15 abnormalities, including heritable imprinting center mutations, cause nonsyndromic Wilms tumor. Nat. Genet. 40, 1329–1334 (2008).
Little, S.E. et al. Frequency and heritability of WT1 mutations in nonsyndromic Wilms' tumor patients: a UK Children's Cancer Study Group Study. J. Clin. Oncol. 22, 4140–4146 (2004).
Mailman, M.D. et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 39, 1181–1186 (2007).
Kim, E., Cho, K.O., Rothschild, A. & Sheng, M. Heteromultimerization and NMDA receptor–clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron 17, 103–113 (1996).
Humbert, P.O. et al. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene 27, 6888–6907 (2008).
Bilder, D., Li, M. & Perrimon, N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113–116 (2000).
Wells, J., Rivera, M.N., Kim, W.J., Starbuck, K. & Haber, D.A. The predominant WT1 isoform (+KTS) encodes a DNA-binding protein targeting the planar cell polarity gene Scribble in renal podocytes. Mol. Cancer Res. 8, 975–985 (2010).
Li, L., Monckton, E.A. & Godbout, R. A role for DEAD box 1 at DNA double-strand breaks. Mol. Cell. Biol. 28, 6413–6425 (2008).
Schaub, R. et al. Array comparative genomic hybridization reveals unbalanced gain of the MYCN region in Wilms tumors. Cancer Genet. Cytogenet. 172, 61–65 (2007).
Squire, J.A. et al. Co-amplification of MYCN and a DEAD box gene (DDX1) in primary neuroblastoma. Oncogene 10, 1417–1422 (1995).
D'Angio, G.J. The National Wilms Tumor Study: a 40 year perspective. Lifetime Data Anal. 13, 463–470 (2007).
Myers, S., Bottolo, L., Freeman, C., McVean, G. & Donnelly, P. A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005).
Acknowledgements
We thank the families and the physicians and nurses that recruited them for their participation in this study, which was funded by the Wellcome Trust Case Control Consortium 3 (WTCCC3) initiative (grant reference 088804/Z/09/Z). We thank P. Donnelly for statistical advice throughout the design and execution of the study. We thank D. Dudakia, J. Bull, R. Linger, B. Ebbs, D. Hughes from Institute of Cancer Research (ICR) and Y. Mistry (from the Children's Cancer and Leukaemia Group (CCLG) tumor bank) for assistance in sample collection, DNA extraction and genotyping. The US samples and data were provided by the Children's Oncology Group (study AREN09B1) supported by the Chair's grant U10 CA98543, SDC grant U10 CA98413 and Human Specimen Banking grant U24 CA114766 from the National Cancer Institute at the US National Institutes of Health. The UK samples were collected through the Factors Associated with Childhood Tumors (FACT) study, which is a CCLG Study (Multicentre Research Ethics Committee (MREC) 05/MRE02/17) and is supported by Cancer Research UK (grant references C8620/A9024 and C8620_A8857). A full list of collaborators is given in the Supplementary Note. The CCRG receives funding from the UK Department of Health, the National Cancer Intelligence Network, the Scottish Government and Children with Cancer UK. The views expressed in this publication are those of the authors and not necessarily of any of these organizations. We acknowledge use of DNA from the British 1958 Birth Cohort DNA collection. We acknowledge use of data from the database of Genotypes and Phenotypes (dbGaP) from the NCBI, US National Library of Medicine. Research support to collect data and develop an application to support High Density SNP Association Analysis of Melanoma (phs000187) was provided by grants 3P50CA093459, 5P50CA097007, 5R01ES011740 and 5R01CA133996 from the US National Institutes of Health. This study also used, in part, data from the National Institute of Neurological Disorders and Stroke (NINDS) dbGaP database from the CIDR:NGRC Parkinson's Disease Study (phs000196). We acknowledge NHS funding to the ICR/Royal Marsden Hospital (RMH)/National Institute for Health Research (NIHR) Specialist Biomedical Cancer Research Centre. I.S. is supported by Michael and Betty Kadoorie Cancer Genetics. C.T. is a Medical Research Council Clinical Research Fellow. This study was conducted at the Institution of Cancer Research, UK.
Author information
Authors and Affiliations
Contributions
N.R. and C.T. designed the study and obtained financial support. M.G., J.H., M.H., J.K., S.L., G.L., M.M., B.M., V.N., J.N., S.P., B.P., M.R., M.S., H.T. and N.R. undertook sample and data collection of UK cases, which was coordinated by A.Z., M.W.-P., K.P.-J., C.A.S. and N.R. A.N., J.D. and P.G. coordinated the transfer of US samples. E.R.P., S.S., R.M.M.-X., S.H., I.S. and A.R. coordinated sample management and replication genotyping and sequencing. D.P. coordinated data transfer and management. C.T. conducted statistical analyses with assistance from D.P. and E.R. C.T., E.R.P. and N.R. wrote the manuscript. N.R. and C.T. oversaw and managed all aspects of the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Table 1–7, Supplementary Figures 1 and 2 and Supplementary Note (PDF 205 kb)
Rights and permissions
About this article
Cite this article
Turnbull, C., Perdeaux, E., Pernet, D. et al. A genome-wide association study identifies susceptibility loci for Wilms tumor. Nat Genet 44, 681–684 (2012). https://doi.org/10.1038/ng.2251
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2251
This article is cited by
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
METTL14 gene polymorphisms decrease Wilms tumor susceptibility in Chinese children
BMC Cancer (2021)
-
Low DLG2 gene expression, a link between 11q-deleted and MYCN-amplified neuroblastoma, causes forced cell cycle progression, and predicts poor patient survival
Cell Communication and Signaling (2020)
-
The genetic changes of Wilms tumour
Nature Reviews Nephrology (2019)
-
Childhood cancer research in Oxford II: The Childhood Cancer Research Group
British Journal of Cancer (2018)