Abstract
A major problem in evolutionary biology is explaining the success of mutualism. Solving this problem requires understanding the level of fidelity between interacting partners. Recent studies have proposed that fungus-growing ants and their fungal cultivars are the products of 'diffuse' coevolution, in which single ant and fungal species are not exclusive to one another. Here we show for ants and associated fungi in the Cyphomyrmex wheeleri species group that each ant species has been exclusively associated with a single fungal cultivar 'species' for millions of years, even though alternative cultivars are readily available, and that rare shifts to new cultivars are associated with ant speciation. Such long-term partner fidelity may have facilitated 'tight' ant–fungus coevolution, and shifts to new fungal cultivars may have had a role in the origin of new ant species.
Similar content being viewed by others
Introduction
Mutualisms occur at all biological levels—from mitochondria within eukaryotic cells to a myriad of species interactions—and they contribute significantly towards increasing complexity in the evolution of life1,2,3,4,5. The long-term stability and success of mutualisms present a challenge for evolutionary theory, for how can such cooperative associations persist, much less proliferate, when selection favours selfishness and conflict4,6,7?
Answering this fundamental question requires understanding of the level of fidelity between interacting partners. Mutualists can be tightly integrated, resulting in coadaptation and cospeciation (that is, speciation in one partner results in simultaneous speciation in the other), or 'diffusely' associated because of frequent host switching. Diffuse coevolution has been proposed to occur in a variety of mutualisms, such as those between figs and fig-wasp pollinators4,8,9, squids and their bioluminescent bacterial symbionts10, and fungus-growing insects and their cultivated fungi11,12,13,14,15,16,17.
All of the 235 described species of fungus-growing ants (tribe Attini) obligately depend on the cultivation of fungus for food18. In return, the ants nourish, protect and disperse their fungal cultivars18. Although all attine ants are obligate symbionts, the cultivated fungi vary from facultative to obligate in their associations with ants. In 'lower' attine fungiculture, the ancestral fungicultural system for attine ants12,19,20, fungal cultivars belong to two distinct, separate lineages ('Clade 1' and 'Clade 2') within the tribe Leucocoprineae. Lower-attine fungi are thought to facultatively associate with ants because the fungi can also live outside the symbiosis12,21. In 'higher' attine fungiculture, however, a highly derived group of fungi descended from a lower-attine fungal ancestor is obligately associated with ants and, as far as is known, cannot live outside the symbiosis13,14,16,17,22,23.
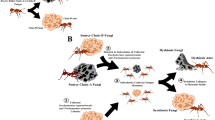
Because cultivar clones are typically propagated vertically from parent to offspring nests via transport by foundress queens, it had been long assumed that the two mutualistic partners evolve by strict coevolution (that is, a fungal species evolves in close association with its host ant species)19,24. However, empirical work over the past two decades has shown that cultivars are frequently transmitted horizontally between different ant colonies (sometimes between colonies of different ant species) and that lower-attine cultivars are closely related to, and in some cases identical to, free-living fungal species, suggesting frequent import of fungi into the symbiosis from free-living sources12,13,14,17,21,23,25,26,27,28,29. This apparent absence of one-to-one host fidelity (that is, long-term association of ant and fungal genotypes), combined with the recent discovery that the lower-attine fungal Clade 2 (Fig. 1) may be evolutionarily much younger than its ant hosts16, has led to a general expectation of low partner fidelity (that is, frequent reassociation of ant and fungal genotypes) in lower-attine fungiculture.
The phylogeny was generated by Bayesian analyses of ITS sequence for 405 taxa (271 unique genotypes). Free-living fungi are indicated by black branches. Ant-associated fungi (red branches) have arisen multiple times and are concentrated in cultivar Clade 1 (shown in blue) and Clade 2 (shown in yellow).
Because of the difficulty of obtaining large samples of both ants and fungi from the same nests across the full geographic range of a species, most previous work on the fidelity of association of attine ants and their fungi involved reconstructing phylogenies of fungi but not of their ant hosts, or of ants and fungi drawn from limited localities12,14,21,26. A complete understanding of the shared evolutionary history of attine ants and their associated fungi, however, requires detailed understanding of the phylogenies of both interacting partners across their geographic ranges30, preferably ant–fungus pairs sampled from the same nests.
It has long been known that two lower attine species in Panama, Cyphomyrmex longiscapus and C. muelleri, cultivate phylogenetically distantly related fungi from Clades 1 and 2, respectively (Fig. 1), even though nests of the two species occur in sympatry and, indeed, may be separated by only a few centimetres in some localities12,26,31,32,33. Based on evidence from morphology and biogeography, it was previously hypothesized that C. muelleri may be derived from an ancestor resembling C. longiscapus s.l. and that the origin of C. muelleri may have coincided with, or even have been driven by, a switch from the Clade 1 to Clade 2 cultivar31. The younger evolutionary age of Clade 2 relative to Clade 116 is consistent with this scenario, as is experimental work demonstrating that Clade 2-cultivating Panamanian C. muelleri ants accept the cultivar of C. longiscapus, whereas C. longiscapus ants do not readily accept the cultivar of C. muelleri33,34, reflecting an asymmetric ancestral/derived host-tolerance pattern also known from insect–plant coevolution35.
These prior observations on the C. longiscapus and C. muelleri symbioses prompted the present investigation of the co-phylogenetic patterns between ants and fungi of these species and their close relatives in the Cyphomyrmex wheeleri group. Here, we reconstruct the relationships of 138 individual ants and 405 fungus-cultivar samples, including ants and fungi taken from the same 88 nests of members of the 'lower' attine C. wheeleri species group (currently consisting of the four species C. wheeleri, C. costatus, C. longiscapus and C. muelleri) drawn from their known ranges in North, Central and South America. Contrary to expectation, we found that each ant species has been exclusively associated with a single fungal cultivar 'species' for millions of years—even though alternative cultivars are readily available—and that rare shifts to new cultivars are associated with ant speciation. These results strongly support a pattern of ant fidelity to particular fungi in this clade of fungus-growing ants.
Results
Global phylogeny of the fungal tribe Leucocoprineae
Consistent with previous studies12,21, our ITS-based phylogeny of the fungal tribe Leucocoprineae (Fig. 1) indicates that associations of fungi with ants have arisen repeatedly, that particular leucocoprineaceous species may be both ant-associated and free-living, and that ant-associated fungi are non-randomly distributed in the phylogeny and concentrated within the 'Clade 1' and 'Clade 2' fungus groups (T-PTP test: nglobal=271, nClade 1=138, nClade 2=69; P<0.001).
Clade 1 and Clade 2 fungal cultivar phylogenies
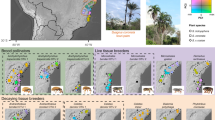
Within 'Clade 1' and 'Clade 2', cultivar associations with C. wheeleri-group ants are concentrated within particular, narrowly defined lineages (Fig. 2a and b). For example, although cultivar clade B (Fig. 2a) includes fungi associated with seven observed fungus-growing ant species, a derived lineage within this clade (darker coloured box) accounts for all associations with Cyphomyrmex cf. longiscapus n. sp. 2 and, within this derived lineage, a single genotype (that is, a single ITS sequence-identical strain) (red arrow), occurring in Ecuador, Panama and Costa Rica, accounts for 64 of 70 such associations. Likewise, although cultivar clade D (Fig. 2a) includes fungi associated with three observed ant species, a derived lineage within that clade (darker coloured box) accounts for 18 of a total of 20 observed associations with C. longiscapus. Finally, cultivar clade G (Fig. 2b) is associated with five observed ant species, but a derived lineage within that clade (darker coloured box) accounts for all observed associations with just two species, C. muelleri and C. cf. costatus sp. 2; within that derived lineage, a single genotype (red arrow), occurring in Ecuador, Panama and Costa Rica, accounts for 49 out of 82 such associations. These patterns indicate that each ant species appears to be exclusively associated with a single species of fungus independent of geography.
(a) Clade 1 and (b) Clade 2. Phylogenies were generated by Bayesian analyses of ITS sequence for 183 and 123 taxa (138 and 69 unique genotypes, respectively). Ant-associated fungi (red branches), designated by associated ant species names and collection IDs, are concentrated in six main clades (A-G, coloured boxes). Within those clades, C. wheeleri group-associated fungi are concentrated in three sublineages (clades B, D and G) and, within those lineages, exclusively (clades B and G) or largely (clade D) in particular sublineages (darker coloured boxes). Red arrows in clades B and G indicate single widespread genotypes (number of ITS sequence-identical strains indicated in parentheses) that account for the great majority (64 of 70 in clade B; 49 of 82 in clade G) of observed associations. Coloured circles indicate posterior probabilities of clades, and small coloured labels indicate geographic localities (see legend). Fungal sequences listed in black were generated for this study; those listed in green result from prior studies.
C. wheeleri group ant phylogeny
The ant phylogeny (Fig. 3) indicates that, in addition to four known and two previously hypothesized cryptic species (C. cf. longiscapus n. sp. 1 and 231), currently circumscribed ant species consist of additional, previously undetected, sibling species. C. wheeleri and C. muelleri are each divided into two distinct geographic species, Texas versus California (K2P-corrected distance=0.11) and Ecuador versus Panama (K2P-corrected distance=0.06), respectively. The geographic species pairs in both C. wheeleri and C. muelleri are old, diverging approximately 5.3 and 5.8 mya, respectively (Fig. 3). C. costatus is also divided into two distinct species-level lineages (K2P-corrected distance=0.10) that diverged approximately 7 mya and that co-occur in Ecuador. C. cf. longiscapus n. sp.1, C. cf. longiscapus n. sp. 2, and C. longiscapus co-occur in Panama.
The phylogeny was generated by Bayesian (shown) and maximum-likelihood analyses of 1,043 bp of mtCOI and 634 bp of nuclear wingless genes from 138 taxa (96 unique genotypes). Large coloured boxes indicate ancestral associations of ants with the cultivar clades shown in Fig. 2, as reconstructed in BayesTraits (all posterior probabilities ≥0.98). Although geographic speciation events within C. wheeleri s.l. and C. muelleri s.l. were not associated with shifts in cultivar association, five observed shifts in cultivar association were consistently associated with ant speciation events, for example, in C. costatus s.l. (sp. 1 versus sp. 2), C. longiscapus s.l. (sp. 1 and sp. 3 versus sp. 2) and at the origin of C. muelleri (C. muelleri s.l. versus C. longiscapus s.l.). Divergence-date estimates from Bayesian relaxed-clock analyses are indicated for major lineages. Coloured circles indicate Bayesian posterior probabilities (above branch) and maximum likelihood bootstrap proportions (below branch); small coloured labels beside taxon names indicate localities; numbers in parentheses refer to the number of samples belonging to a given ant genotype; and names in non-bold type indicate ant species for which fungal cultivars were not available. Ant heads from top to bottom: C. cf. costatus sp. 2, C. wheeleri sp. 2, C. cf. longiscapus sp. 2, C. longiscapus sp. 3 (syntype specimen) and C. muelleri sp. 2.
Discussion
Contrary to expectation, since their origins, all C. wheeleri group ant species have been tightly associated with narrowly defined fungal groups (Fig. 3) (BayesTraits ancestral-state posterior probabilities [PP] ≥0.98). C. cf. costatus sp. 1, occurring in Ecuador, is entirely associated with cultivar clade B, whereas C. cf. costatus sp. 2, occurring in Ecuador, Costa Rica and Panama, is entirely associated with cultivar clade G. C. cf. longiscapus n. sp. 1 is associated with cultivar clade D, C. cf. longiscapus n. sp. 2 is associated with fungal cultivar clade B, C. longiscapus is associated with cultivar clade D and both C. wheeleri sp. 1 and sp. 2 are associated with cultivar clade A. With a single exception (C. muelleri sp. 2 AGH020526-03), both C. muelleri sp. 1 and sp. 2 are associated with cultivar clade G. Data on the exception, C. muelleri sp. 2 AGH020526-03, were included in BayesTraits ancestral-state analyses and in ParaFit analyses of coevolution and did not alter the respective conclusions that the ancestor of C. muelleri s.l. cultivated a clade G fungus (PP=1.0) and that significant ant–fungus coevolution has occurred between clade G fungi and their associated ants (P<0.05). In short, although not all observed ant speciation events are associated with shifts in cultivar association, every observed shift in cultivar association is associated with an ant speciation event.
Taken together, these results strongly support a pattern of ant fidelity to particular fungi that is independent of geography and stable over evolutionary time spans of several millions of years. For example, C. wheeleri s.l. consists of two distinct geographic species that have both remained associated with cultivar clade A for approximately 5 million years. Likewise, C. muelleri s.l. includes two distinct cryptic sibling species that have both been associated with cultivar clade G for approximately 6 million years (Fig. 3). Whereas these examples represent cases in which ant lineages have diverged while remaining faithful to the same cultivar, observed shifts to new fungal cultivars have invariably been associated with ant speciation1, as discussed above. For example, C. cf. costatus sp. 1 is associated with cultivar clade B, whereas its sister species is associated with cultivar clade G (Fig. 3), representing a shift not only between fungal species but also between Clade 1 and Clade 2 fungi that occurred approximately 7 million years ago. Additional cultivar shifts correlated with ant-speciation events occurred at the origins of C. wheeleri s.l., C. cf. longiscapus n. sp. 1, C. cf. longiscapus n. sp. 2, C. longiscapus and C. muelleri, all occurring 5.3 to 6.9 million years ago. Although this pattern is consistent with a prior hypothesis that shifts in fungal cultivar association may have driven speciation in ants of the C. wheeleri group31, it is also consistent with alternative hypotheses, including those in which ant speciation preceded cultivar shifts. It should also be noted that even though the combined nuclear and mitochondrial K2P-corrected distances between C. wheeleri sp. 1 and 2 and C. muelleri sp. 1 and 2 are quite large, it remains possible that one or both of these two purely geographic, non-sympatric species pairs are the highly divergent endpoints of conspecific population clines. This alternative, however, only strengthens our results because, under that scenario, every case of speciation in the C. wheeleri group is then correlated with an ancestral shift in cultivar association.
This historical pattern of long-term ant-fungus fidelity for several million years provides the opportunity for coevolution, in which evolutionary modification in an ant host can prompt modification in its fungal symbiont, and vice versa. Because cultivar lineages are associated with multiple ant species, the C. wheeleri-group ant–fungus mutualism cannot be characterized as strict 'one-to-one' coevolution, but neither can it be characterized as broadly diffuse, 'one-to-many,' coevolution. Instead, it might best be characterized as narrowly diffuse, 'one-to-few' coevolution. The C. wheeleri ant clade is closely related to the higher Attini20, which includes the leaf-cutting ants whose fungal cultivars are highly derived, obligate symbionts that exhibit coevolved modifications16,20,22. If C. wheeleri-group ant–fungus associations are more specific than those found in other lower Attini, they may represent an intermediate condition linking the broadly diffuse associations found in other lower-attine ants with the narrow associations that led to the evolution of higher-attine fungiculture.
Methods
Samples and DNA preparation
We conducted phylogenetic analyses of C. wheeleri group ants and their associated fungi based on DNA sequence data obtained from a total of 138 ant and 193 fungal samples from Panama, Ecuador, Costa Rica and the United States (Supplementary Table S1). Our data included a subset of 88 nests that yielded exemplars of both ants and fungi. For fungi, we combined 193 fungal DNA sequences obtained for this study with 212 fungal DNA sequences from prior studies12,21 to construct a global phylogeny of 405 leucocoprineacous fungi (reduced to 271 when all but one exemplar of each group of redundant genotypes were excluded; Supplementary Table S2). These samples were in most cases preserved in ethanol in the field; some fungi were cultured from gardens of laboratory nests and their mycelium was lyophilized and preserved at −80 °C for later study. Ant vouchers and fungal samples are deposited in the USNM Department of Entomology collections. Information detailing extraction, PCR and sequencing methods can be found in the Supplementary Methods.
Alignments and data matrices
Ants: For this study we generated sequence data for 138 individual ants consisting of a 1,043-bp COI protein-coding fragment, two wg protein-coding fragments totalling 316 bp and a variable-length wg intron consisting of 220–318 bp. Sequences were edited in Sequencher (Lasergene version 7.0; DNASTAR, Madison, WI) and aligned using ClustalW36 and by eye. After eliminating all but one representative of each unique genotype and one taxon judged to contain a COI pseudogene, and after excluding 44 bp of autapomorphic indel regions occurring in the wg intron, our ant data set consisted of 96 ant taxa and 1720 nucleotide sites. Because 32 taxa contained one or more missing fragments, for each treatment two phylogenetic analyses were conducted, one with the full 96-taxon data set and one with the 64-taxon data set in which there were no missing fragments. To check for possible gene tree/species tree discordance problems, single-gene analyses were also conducted utilizing data matrices consisting of, for COI, 96 taxa and 1,043 nucleotide characters and, for wg, 68 taxa and 633 nucleotide characters. Fungi: For this study we generated ITS sequence data for 193 ant garden fungi for a variable-length fragment (∼700–750 bp) of the nuclear ITS region. After eliminating all but one representative of each unique genotype, our fungal data set consisted of 59 exemplar taxa, which we analysed in three separate alignments: global, Clade 1 and Clade 2. Global analyses utilized a data matrix in which the 59 new sequences were combined with 212 leucocoprineaceous sequences generated for previous studies, producing a data matrix of 271 fungal taxa and 721 nucleotide sites. Clade 1 analyses utilized a data matrix of 138 taxa (out of a total of 183 Clade 1 taxa), including all Clade 1 fungi known to be ant-associated, and 838 nucleotide sites. Clade 2 analyses utilized a data matrix of 69 taxa and 950 nucleotide sites. All alignments were produced in the program MAFFT v6.814b37 using the E-INS-I algorithm, a 200PAM/k=2 scoring matrix, a gap opening penalty of 1.53, and an offset value of zero.
Data partitioning
Ants: Based on genes and on the variability of codon–position sites within each gene, and based on the results of Ward et al.38, we partitioned the ant data set into six partitions: COI first positions, COI second positions, COI third positions, wg first and second positions, wg third positions and the wg intron. Fungi: In global analyses, the fungal ITS region was analysed as a single unpartitioned unit. For Clade 1 and Clade 2 analyses, we partitioned the fungal ITS region into two partitions, a 'rapidly evolving' partition and a 'conserved' partition. In all cases, best-fit models of sequence evolution were selected for each partition under the Akaike information criterion as calculated in ModelTest v3.739.
Bayesian phylogenetic inference
All ant and fungal data sets were analysed under Bayesian criteria as implemented in MrBayes v3.1. 240 with nucmodel=4by4 and samplefreq=500 (ants) or 100 (fungi). All parameters, including branch-length rate multipliers, were unlinked across partitions except branch lengths and topology. All analyses were carried out using parallel processing (one chain per CPU) with 8 chains per run and 2 runs per analysis (nruns=2) and consisted of 10 million generations.
Maximum likelihood analyses
All ant data sets were analysed under maximum likelihood criteria as implemented in Garli 0.97. r73741,42 using parallel processing. ML bootstrap analyses consisted of 1000 pseudoreplicates, deviating from default settings as follows: genthreshfortopoterm=5000; scorethreshforterm=0.10; startoptprec=0.5; minoptprec=0.01; numberofprecreductions=1; treerejectionthreshold=20.0; topoweight=0.01; brlenweight=0.002. ML 'best tree' analyses consisted of 100 searches, deviating from the default settings as follows: topoweight=0.01; brlenweight=0.002.
T-PTP tests
The correlation with phylogeny of ant-associated vs. free-living fungi was tested for the global (n=271), Clade 1 (n=138), and Clade 2 (n=69) fungal phylogenies using the T-PTP test43 with 1000 iterations as implemented in PAUP* v.4.0a118.44 In each case, the significance of the parsimony-based length difference between the observed tree and the tree in which ant-associated fungi were constrained to be monophyletic was assessed by comparing it to the distribution of such length differences calculated from 1,000 replicates in which ant associations were randomized across fungal taxa. For all three data sets, the observed length difference was consistently less than the length differences for the permuted data, that is, the observed length difference was significant at P<0.001.
Test for coevolution
The permutation-based ParaFit test for significant global coevolution between 'parasites' (attine ants) and 'hosts' (leucocoprineaceous fungi)45 was applied to a reduced data set of 99 ants and 90 fungi, that is, to the largest overlapping subset of ants and fungi for which we had sequence data and host associations and, for ants, for which we had sufficient sequence data to calculate pairwise distances. Distances were calculated for both ants and fungi in PAUP* v.4.0a11844 under the GTR+I+G model using maximum-likelihood values for the proportion of sites invariant and gamma alpha calculated in PAUP on maximum-likelihood topologies identified by GARLI-PART 0.97.r73741. Distances were converted into principal coordinate matrices46 in DistPCoA47 with negative eigenvalues removed.
The principal-coordinate-transformed distance data were analysed in ParaFit using 999 permutations, resulting in significant support for a hypothesis of global coevolution between Cyphomyrmex wheeleri-group ants and their fungal cultivars (P=0.001), as well as for ant-fungus coevolution in fungal clades A, B, D, F and G (P<0.05) (Figs 2 and 3).
Ancestral state reconstruction
We inferred the ancestral fungal lineage for each ant species using the Bayesian method48 implemented by BayesMultiState in the computer package BayesTraits (www.evolution.rdg.ac.uk). We coded each fungal lineage within the Cyphomyrmex wheeleri group (cultivars A, B, D, F, G) as a separate state; in cases where the fungal association was unknown for a particular ant specimen, the state was coded as unknown. We estimated Bayesian posterior distributions for each state using MCMC on a set of 1,000 trees selected randomly without replacement from the total set of post-burnin trees obtained from the MrBayes analyses. As suggested in the program manual, exploratory analyses were conducted to determine the appropriate range for hyperpriors and to select a ratedev value that produced an acceptance rate between 20 and 40%. We used hpall (0, 10) and ratedev=20 for the results reported below; alternative distributions and values produced virtually identical results. The MCMC chain was run for 100 million generations with a burnin of 10 million and sampled every 10,000 generations. The following fungal cultivar lineages were reconstructed at the crown-group node for each ant species: C. strigatus group n. sp., cultivar F with PP=0.99; C. faunulus, cultivar F with PP=1.0; C. costatus sp.1, cultivar B with PP=1.0; C. costatus sp.2, cultivar G with PP=1.0; C. wheeleri, cultivar A with PP=0.98; C. longiscapus sp.1, cultivar D with PP=1.0; C. longiscapus sp.2, cultivar B with PP=1.0; C. longiscapus sp.3, cultivar D with PP=1.0; C. muelleri, cultivar G with PP=1.0.
Divergence dating
Divergence times for ant lineages within the Cyphomyrmex wheeleri group were inferred under a Bayesian framework using BEAST v.1. 6.1 by employing an uncorrelated lognormal-relaxed clock model49,50. We used the same nucleotide-partitioning scheme and substitution models as detailed above. Substitution models were unlinked and clock and tree models linked among partitions. A Yule speciation process with a random starting tree was used for the tree prior. We assigned a priori age distributions to two nodes. The root node was assigned a lognormal prior of log(mean)=1.6, log (s.d.)=1.0, offset=15 (equivalent to a minimum bound of 15 Ma, median value of 20 Ma and a 95% quantile of 40 Ma) based on the presence of Cyphomyrmex maya and C. taino, both members of the C. rimosus group, in Dominican amber20,51. The crown group of the C. wheeleri group was also assigned a normal prior of mean=20, s.d.=5 (equivalent to a median of 20 Ma and a 95% confidence interval of 1.8–18.2 Ma) based on prior divergence dates from a largely independent data set20. Elimination of this second prior, which is not directly linked to the fossil record, altered resulting ages by less than 1 Ma. MCMC searches were conducted for 100,000,000 generations with the first 20,000,000 generations discarded as burn-in. Trees and other parameter values were saved every 10,000 generations. Convergence and stationarity were assessed with Tracer v.1.552 using ESS scores, log likelihood values and the consistency of results between multiple runs. The results from two independent runs were combined and visualized using FIGTREE v.1.3.153.
Additional information
Accession codes: The sequence data have been deposited in GenBank NCBI database under accession codes JQ617314 to JQ617746 listed in Supplementary Tables S1 and S2.
How to cite this article: Mehdiabadi, N. J. et al. Symbiont fidelity and the origin of species in fungus-growing ants. Nat. Commun. 3:840 doi: 10.1038/ncomms1844 (2012).
References
Wallin, I. E. Symbionticism and the Origin of Species (Waverly Press, 1927).
Thompson, J. N. The Coevolutionary Process (Univ. of Chicago Press, 1994).
Maynard Smith, J. & Szathmáry, E. The Major Transitions in Evolution (Oxford Univ. Press, 1995).
Herre, E. A., Knowlton, N., Mueller, U. G. & Rehner, S. A. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol. Evol. 14, 49–53 (1999).
Bourke, A. F. G. Principles of Social Evolution (Oxford Univ. Press, 2011).
Sachs, J. L., Mueller, U. G., Wilcox, T. P. & Bull, J. J. Evolution of cooperation. Q. Rev. Biol. 79, 135–160 (2004).
Foster, K. R. & Wenseleers, T. A general model for the evolution of mutualisms. J. Evol. Biol. 19, 1283–1293 (2006).
Machado, C. A., Robbins, N., Gilbert, M. T. P. & Herre, E. A. Critical review of host specificity and its coevolutionary implications in the fig/fig-wasp mutualism. Proc. Natl Acad. Sci. USA 102, 6558–6565 (2005).
Jackson, A. P., Machado, C. A., Robbins, N. & Herre, E. A. Multi-locus phylogenetic analysis of neotropical figs does not support co-speciation with the pollinators: the importance of systematic scale in fig/wasp cophylogenetic studies. Symbiosis 45, 57 (2008).
Dunlap, P. V. et al. Phylogenetic analysis of host symbiont specificity and codivergence in bioluminescent symbioses. Cladistics 23, 507 (2007).
Nobre, T., Fernandes, C., Boomsma, J. J., Korb, J. & Aanen, D. K. Farming termites determine the genetic population structure of Termitomyces fungal symbionts. Mol. Ecol. 20, 2023–2033 (2011).
Mueller, U. G., Rehner, S. A. & Schultz, T. R. The evolution of agriculture in ants. Science 281, 2034–2038 (1998).
Mikheyev, A. S., Mueller, U. G. & Abbott, P. Cryptic sex and many-to-one co-evolution in the fungus-growing ant symbiosis. Proc. Natl Acad. Sci. USA 103, 10702–10706 (2006).
Mikheyev, A. S., Mueller, U. G. & Boomsma, J. J. Population genetic signatures of diffuse coevolution between Panamanian leaf-cutter ants and their cultivar fungi. Mol. Ecol. 16, 209–216 (2007).
Mikheyev, A. S., Vo, T. & Mueller, U. G. Phylogeography of post-Pleistocene population expansion in a fungus-gardening ant and its microbial mutualists. Mol. Ecol. 17, 4480 (2008).
Mikheyev, A. S., Mueller, U. G. & Abbott, P. Comparative dating of attine ant and lepiotaceous cultivar phylogenies reveals coevolutionary synchrony and discord. Am. Nat. 175, E126–E133 (2010).
Mueller, U. G., Mikheyev, A. S., Solomon, S. E. & Cooper, M. Frontier mutualism: Co-evolutionary patterns at the northern range limit of the leafcutter ant-fungus symbiosis. Proc. R. Soc. Lond. B 278, 3050–3059 (2011).
Mehdiabadi, N. J. & Schultz, T. R. Natural history and phylogeny of the fungus-farming ants (Hymenoptera: Formicidae: Myrmicinae: Attini). Myr. News 13, 37–55 (2010).
Chapela, I. H., Rehner, S. A., Schultz, T. R. & Mueller, U. G. Evolutionary history of the symbiosis between fungus-growing ants and their fungi. Science 266, 1691–1694 (1994).
Schultz, T. R. & Brady, S. G. Major evolutionary transitions in ant agriculture. Proc. Natl Acad. Sci. USA 105, 5435–5440 (2008).
Vo, T. L., Mikheyev, A. S. & Mueller, U. G. Free-living fungal symbionts (Lepiotaceae) of fungus-growing ants (Attini: Formicidae). Mycologia 101, 206–210 (2009).
Schultz, T. R., Mueller, U. G., Currie, C. R. & Rehner, S. A. in Ecological and Evolutionary Advances in Insect-Fungal Associations (eds Vega, F. & Blackwell, M.) Ch. 7, pp. 149–190 (Oxford Univ. Press, 2005).
Silva-Pinhati, A. C. O. et al. Low variation in ribosomal DNA and internal transcribed spacers of the symbiotic fungi of leaf-cutting ants (Attini: Formicidae). Braz. J Med. Biol. Res. 37, 1463–1472 (2004).
Hinkle, G., Wetterer, J. K., Schultz, T. R. & Sogin, M. L. Phylogeny of the attine ant fungi based on analysis of small subunit ribosomal RNA gene sequences. Science 266, 1695–1697 (1994).
Bot, A. N. M., Rehner, S. A. & Boomsma, J. J. Partial incompatibility between ants and symbiotic fungi in two sympatric species of Acromyrmex leaf-cutting ants. Evolution 55, 1980–1991 (2001).
Green, A. M., Mueller, U. G. & Adams, R. M. M. Extensive exchange of fungal cultivars between sympatric species of fungus-growing ants. Mol. Ecol. 11, 191–195 (2002).
Mueller, U. G., Schultz, T. R., Currie, C. R., Adams, R. M. M. & Malloch, D. The origin of the attine ant-fungus mutualism. Q. Rev. Biol. 76, 169–197 (2001).
Munkacsi, A. B. et al. Convergent coevolution in the domestication of coral mushrooms by fungus-growing ants. Proc. R. Soc. Lond. B 271, 1777–1782 (2004).
Dentinger, B. T. M., Lodge, D. J., Munkacsi, A. B., Desjardin, D. E. & McLaughlin, D. J. Phylogenetic placement of an unusual coral mushroom challenges the classic hypothesis of strict coevolution in the Apterostigma pilosum group ant-fungus mutualism. Evolution 63, 2172–2178 (2009).
Thompson, J. N. The Geographic Mosaic of Coevolution (Univ. of Chicago Press, 2005).
Schultz, T. R. et al. Cryptic speciation in the fungus-growing ants Cyphomyrmex longiscapus Weber and Cyphomyrmex muelleri Schultz and Solomon, new species (Formicidae: Attini). Insectes Sociaux 49, 331–343 (2002).
Mueller, U. G. & Wcislo, W. T. Nesting biology of the fungus-growing ant Cyphomyrmex longiscapus Weber (Attini, Formicidae). Insectes Sociaux 45, 181–189 (1998).
Mehdiabadi, N. J., Hughes, B. & Mueller, U. G. Cooperation, conflict, and coevolution in the attine ant-fungus symbiosis. Behav. Ecol. 17, 291–296 (2006).
Mueller, U. G., Poulin, J. & Adams, R. M. M. Symbiont choice in a fungus-growing ant (Attini, Formicidae). Behav. Ecol. 15, 357–364 (2004).
Winkler, I. S. & Mitter, C. in Specialization, Speciation, and Radiation: The Evolutionary Biology of Herbivorous Insects (ed Tilmon, K. J.) Ch. 18, 240–263 (Univ. California Press, 2008).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Ward, P. S., Brady, S. G., Fisher, B. L. & Schultz, T. R. Phylogeny and biogeography of dolichoderine ants: effects of data partitioning and relict taxa on historical inference. Syst. Biol. 59, 342–362 (2010).
Posada, D. & Crandall, K. A. Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818 (1998).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Zwickl, D. J. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion Ph.D. thesis, The University of Texas at Austin (2006).
Zwickl, D. J. GARLI-part v0.97, http://garli.googlecode.com (2010).
Faith, D. P. Cladistic permutation tests for monophyly and nonmonophyly. Syst. Zool. 40, 366–375 (1991).
Swofford, D. L. PAUP*: Phylogenetic Analysis using Parsimony (*and other methods) Version 4, (Sinauer Associates, Sunderland, MA, 2002).
Legendre, P., Desdevises, Y. & Bazin, E. A Statistical test for host-parasite coevolution. Syst. Biol. 51, 217–234 (2002).
Gower, J. C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53, 325–338 (1966).
Legendre, P. & Anderson, M. J. Program DistPCoA (Département de sciences biologiques, Université de Montréal, Canada, 1998).
Pagel, M., Meade, A. & Barker, D. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673–684 (2004).
Drummond, A. J., Ho, S. Y. W., Phillips, M. J. & Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biology 4, e88 (2006).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7, 214 (2007).
de Andrade, M. L. First descriptions of two new amber species of Cyphomyrmex from Mexico and the Dominican Republic (Hymenoptera: Formicidae). Beiträge zur Entomologie 53, 131–139 (2003).
Rambaut, A. & Drummond, A. J. Tracer v1.5, http://tree.bio.ed.ac.uk/software/tracer/ (2009).
Rambaut, A. Figtree v.1.3.1, http://tree.bio.ed.ac.uk/software/figtree/ (2010).
Acknowledgements
We thank Lee Weigt and other members of the Laboratories of Analytical Biology (NMNH) for their help and support throughout this project. We also thank Rachelle Adams, Donat Agosti, Stefan Cover, Cameron Currie, Nicole Gerardo, Lacey Loudermilk, Alexander Mikheyev, Shauna Price, Kaci Richardson, Andre Rodrigues, Jarrod Scott, Adam Smith, Heraldo Vasconcelos, Santiago Villamarin and Tanya Vo for collecting colonies. We thank Patrick Abbot, Jerry Coyne and Phil Ward for improving earlier versions of the manuscript and Paul Armstrong, Faridah Nor Dahlan, Jeffrey Hunt, Heather Ishak, Matthew Kweskin, Eugenia Okonski, Steve Rehner, Jeffrey Sosa-Calvo, Phil Ward, Hong Zhao and members of the Mueller Lab and the Schultz Lab for help and discussion. Funding was provided by a Smithsonian Postdoctoral Fellowship (NJM), a Smithsonian Scholarly Studies grant (TRS, SGB), NMNH Laboratories of Analytical Biology (NJM, Lee Weigt), and NSF grants DEB-0949689 (TRS, NJM, UGM), DEB-0110073 (UGM, TRS), DEB-0206372 (AGH, UGM), IOS-0920138 (UGM) and EF-0431330 (SGB, TRS).
Author information
Authors and Affiliations
Contributions
N.J.M., U.G.M., and T.R.S. designed experiments; U.G.M., A.G.H., and T.R.S. collected samples; T.R.S. identified ant specimens; N.J.M. performed experiments; N.J.M, S.G.B., and T.R.S. analysed the data; N.J.M. and T.R.S. wrote the manuscript; U.G.M., S.G.B., and A.G.H. commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Tables S1-S2, Supplementary Methods and Supplementary References (PDF 341 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Mehdiabadi, N., Mueller, U., Brady, S. et al. Symbiont fidelity and the origin of species in fungus-growing ants. Nat Commun 3, 840 (2012). https://doi.org/10.1038/ncomms1844
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms1844
This article is cited by
-
Ant genotype, but not genotype of cultivated fungi, predicts queen acceptance in the asexual fungus-farming ant Mycocepurus smithii (Hymenoptera: Formicidae)
Behavioral Ecology and Sociobiology (2023)
-
Garden microbiomes of Apterostigma dentigerum and Apterostigma pilosum fungus-growing ants (Hymenoptera: Formicidae)
Journal of Microbiology (2019)
-
Symbiont-Mediated Host-Parasite Dynamics in a Fungus-Gardening Ant
Microbial Ecology (2018)
-
Context is everything: mapping Cyphomyrmex-derived compounds to the fungus-growing ant phylogeny
Chemoecology (2018)
-
Biology of the relict fungus-farming ant Apterostigma megacephala Lattke, including descriptions of the male, gyne, and larva
Insectes Sociaux (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.