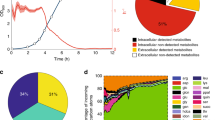
Abstract
Absolute metabolite concentrations are critical to a quantitative understanding of cellular metabolism, as concentrations impact both the free energies and rates of metabolic reactions. Here we use LC-MS/MS to quantify more than 100 metabolite concentrations in aerobic, exponentially growing Escherichia coli with glucose, glycerol or acetate as the carbon source. The total observed intracellular metabolite pool was approximately 300 mM. A small number of metabolites dominate the metabolome on a molar basis, with glutamate being the most abundant. Metabolite concentration exceeds Km for most substrate-enzyme pairs. An exception is lower glycolysis, where concentrations of intermediates are near the Km of their consuming enzymes and all reactions are near equilibrium. This may facilitate efficient flux reversibility given thermodynamic and osmotic constraints. The data and analyses presented here highlight the ability to identify organizing metabolic principles from systems-level absolute metabolite concentration data.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bajad, S.U. et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A 1125, 76–88 (2006).
Coulier, L. et al. Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 78, 6573–6582 (2006).
Luo, B., Groenke, K., Takors, R., Wandrey, C. & Oldiges, M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J. Chromatogr. A 1147, 153–164 (2007).
Tu, B.P. et al. Cyclic changes in metabolic state during the life of a yeast cell. Proc. Natl. Acad. Sci. USA 104, 16886–16891 (2007).
Brauer, M.J. et al. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. USA 103, 19302–19307 (2006).
Oldiges, M., Kunze, M., Degenring, D., Sprenger, G.A. & Takors, R. Stimulation, monitoring, and analysis of pathway dynamics by metabolic profiling in the aromatic amino acid pathway. Biotechnol. Prog. 20, 1623–1633 (2004).
Villas-Bôas, S.G. & Bruheim, P. Cold glycerol-saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal. Biochem. 370, 87–97 (2007).
Mashego, M.R. et al. MIRACLE: mass isotopomer ratio analysis of U-13C-labeled extracts. A new method for accurate quantification of changes in concentrations of intracellular metabolites. Biotechnol. Bioeng. 85, 620–628 (2004).
Seifar, R.M. et al. Quantitative analysis of metabolites in complex biological samples using ion-pair reversed-phase liquid chromatography-isotope dilution tandem mass spectrometry. J. Chromatogr. A 1187, 103–110 (2008).
Wu, L. et al. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal. Biochem. 336, 164–171 (2005).
Zamboni, N., Kummel, A. & Heinemann, M. AnNET: a tool for network-embedded thermodynamic analysis of quantitative metabolome data. BMC Bioinformatics 9, 199 (2008).
Kümmel, A., Panke, S. & Heinemann, M. Systematic assignment of thermodynamic constraints in metabolic network models. BMC Bioinformatics 7, 512 (2006).
Kümmel, A., Panke, S. & Heinemann, M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 2, 2006.0034 (2006).
Henry, C.S., Broadbelt, L.J. & Hatzimanikatis, V. Thermodynamics-based metabolic flux analysis. Biophys. J. 92, 1792–1805 (2007).
Hoppe, A., Hoffmann, S. & Holzhutter, H.G. Including metabolite concentrations into flux balance analysis: thermodynamic realizability as a constraint on flux distributions in metabolic networks. BMC Syst. Biol. 1, 23 (2007).
Yuan, J., Bennett, B.D. & Rabinowitz, J.D. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat. Protoc. 3, 1328–1340 (2008).
Yuan, J., Fowler, W.U., Kimball, E., Lu, W. & Rabinowitz, J.D. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat. Chem. Biol. 2, 529–530 (2006).
Easterby, J.S. The effect of feedback on pathway transient response. Biochem. J. 233, 871–875 (1986).
Jamshidi, N. & Palsson, B.O. Formulating genome-scale kinetic models in the post-genome era. Mol. Syst. Biol. 4, 171 (2008).
Gutenkunst, R.N. et al. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 3, 1871–1878 (2007).
Piazza, M., Feng, X.J., Rabinowitz, J.D. & Rabitz, H. Diverse metabolic model parameters generate similar methionine cycle dynamics. J. Theor. Biol. 251, 628–639 (2008).
Karp, P.D. et al. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 35, 7577–7590 (2007).
Ibarra, R.U., Edwards, J.S. & Palsson, B.O. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420, 186–189 (2002).
Fischer, E. & Sauer, U. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270, 880–891 (2003).
Birkemeyer, C., Luedemann, A., Wagner, C., Erban, A. & Kopka, J. Metabolome analysis: the potential of in vivo labeling with stable isotopes for metabolite profiling. Trends Biotechnol. 23, 28–33 (2005).
Bennett, B.D., Yuan, J., Kimball, E.H. & Rabinowitz, J.D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 3, 1299–1311 (2008).
Kimball, E. & Rabinowitz, J.D. Identifying decomposition products in extracts of cellular metabolites. Anal. Biochem. 358, 273–280 (2006).
Rabinowitz, J.D. & Kimball, E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 79, 6167–6173 (2007).
Edgar, J.R. & Bell, R.M. Biosynthesis in Escherichia coli fo sn-glycerol 3-phosphate, a precursor of phospholipid. J. Biol. Chem. 253, 6348–6353 (1978).
Lin, E.C. Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 30, 535–578 (1976).
Asnis, R.E. & Brodie, A.F. A glycerol dehydrogenase from Escherichia coli. J. Biol. Chem. 203, 153–159 (1953).
Truniger, V. & Boos, W. Mapping and cloning of gldA, the structural gene of the Escherichia coli glycerol dehydrogenase. J. Bacteriol. 176, 1796–1800 (1994).
Alberty, R.A. Standard Gibbs free energy, enthalpy, and entropy changes as a function of pH and pMg for several reactions involving adenosine phosphates. J. Biol. Chem. 244, 3290–3302 (1969).
Maskow, T. & von Stockar, U. How reliable are thermodynamic feasibility statements of biochemical pathways? Biotechnol. Bioeng. 92, 223–230 (2005).
Shikama, K. Standard free energy maps for the hydrolysis of ATP as a function of pH, pMg and pCa. Arch. Biochem. Biophys. 147, 311–317 (1971).
Shikama, K. & Nakamura, K.I. Standard free energy maps for the hydrolysis of ATP as a function of pH and metal ion concentration: comparison of metal ions. Arch. Biochem. Biophys. 157, 457–463 (1973).
Neidhardt, F. et al. Escherichia coli and Salmonella typhimurium, Vol. 1 (American Society for Microbiology, Washington, DC, 1987).
Ikeda, T.P., Shauger, A.E. & Kustu, S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259, 589–607 (1996).
McLaggan, D., Naprstek, J., Buurman, E.T. & Epstein, W. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J. Biol. Chem. 269, 1911–1917 (1994).
Beg, Q.K. et al. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc. Natl. Acad. Sci. USA 104, 12663–12668 (2007).
Powell, J.T. & Morrison, J.F. The purification and properties of the aspartate aminotransferase and aromatic-amino-acid aminotransferase from Escherichia coli. Eur. J. Biochem. 87, 391–400 (1978).
Miller, R.E. & Stadtman, E.R. Glutamate synthase from Escherichia coli. An iron-sulfide flavoprotein. J. Biol. Chem. 247, 7407–7419 (1972).
Freedberg, W.B., Kistler, W.S. & Lin, E.C. Lethal synthesis of methylglyoxal by Escherichia coli during unregulated glycerol metabolism. J. Bacteriol. 108, 137–144 (1971).
Benov, L., Beema, A.F. & Sequeira, F. Triosephosphates are toxic to superoxide dismutase-deficient Escherichia coli. Biochim. Biophys. Acta 1622, 128–132 (2003).
Soupene, E. et al. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185, 5611–5626 (2003).
Gutnick, D., Calvo, J.M., Klopotowski, T. & Ames, B.N. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhirium LT-2. J. Bacteriol. 100, 215–219 (1969).
Rosenberg, H., Russell, L.M., Jacomb, P.A. & Chegwidden, K. Phosphate exchange in the pit transport system in Escherichia coli. J. Bacteriol. 149, 123–130 (1982).
Acknowledgements
We are indebted to B. Cravatt for the suggestion that metabolite concentrations be compared to the Km of consuming enzymes. We thank the US National Science Foundation (CAREER Award), the Arnold and Mabel Beckman Foundation, the US National Institutes of Health (Center for Quantitative Biology grant P50 GM071508) and the American Heart Association for their financial support.
Author information
Authors and Affiliations
Contributions
B.D.B. performed experiments, analyzed data and wrote the paper. E.H.K. and M.G. performed experiments. R.O. and S.J.V.D. conducted the TMFA. J.D.R. analyzed data and wrote the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1, Supplementary Tables 1–8 and Supplementary Methods (PDF 363 kb)
Rights and permissions
About this article
Cite this article
Bennett, B., Kimball, E., Gao, M. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5, 593–599 (2009). https://doi.org/10.1038/nchembio.186
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.186
This article is cited by
-
Lipid membrane remodeling and metabolic response during isobutanol and ethanol exposure in Zymomonas mobilis
Biotechnology for Biofuels and Bioproducts (2024)
-
Systematic engineering pinpoints a versatile strategy for the expression of functional cytochrome P450 enzymes in Escherichia coli cell factories
Microbial Cell Factories (2023)
-
Network-wide thermodynamic constraints shape NAD(P)H cofactor specificity of biochemical reactions
Nature Communications (2023)
-
Stress-induced metabolic exchanges between complementary bacterial types underly a dynamic mechanism of inter-species stress resistance
Nature Communications (2023)
-
Learning perturbation-inducible cell states from observability analysis of transcriptome dynamics
Nature Communications (2023)