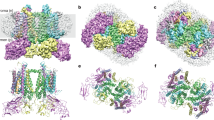
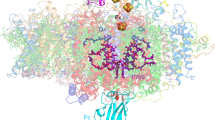
Abstract
During photosynthesis, the plant photosystem II core complex receives excitation energy from the peripheral light-harvesting complex II (LHCII). The pathways along which excitation energy is transferred between them, and their assembly mechanisms, remain to be deciphered through high-resolution structural studies. Here we report the structure of a 1.1-megadalton spinach photosystem II–LHCII supercomplex solved at 3.2 Å resolution through single-particle cryo-electron microscopy. The structure reveals a homodimeric supramolecular system in which each monomer contains 25 protein subunits, 105 chlorophylls, 28 carotenoids and other cofactors. Three extrinsic subunits (PsbO, PsbP and PsbQ), which are essential for optimal oxygen-evolving activity of photosystem II, form a triangular crown that shields the Mn4CaO5-binding domains of CP43 and D1. One major trimeric and two minor monomeric LHCIIs associate with each core-complex monomer, and the antenna–core interactions are reinforced by three small intrinsic subunits (PsbW, PsbH and PsbZ). By analysing the closely connected interfacial chlorophylls, we have obtained detailed insights into the energy-transfer pathways between the antenna and core complexes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
The cryo-EM map of the spinach PSII–LHCII supercomplex has been deposited in the Electron Microscopy Data Bank with accession code EMD-6617. The corresponding structure model has been deposited in the Protein Data Bank under accession code 3JCU.
Change history
01 June 2016
The received date year was corrected from 2015 to 2016.
References
Nelson, N. & Junge, W. Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 84, 659–683 (2015)
Shen, J.-R. The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 66, 23–48 (2015)
Vinyard, D. J., Ananyev, G. M. & Dismukes, G. C. Photosystem II: The reaction center of oxygenic photosynthesis. Annu. Rev. Biochem. 82, 577–606 (2013)
Pan, X., Liu, Z., Li, M. & Chang, W. Architecture and function of plant light-harvesting complexes II. Curr. Opin. Struct. Biol. 23, 515–525 (2013)
Barros, T. & Kühlbrandt, W. Crystallisation, structure and function of plant light-harvesting Complex II. Biochim. Biophys. Acta 1787, 753–772 (2009)
Croce, R. & van Amerongen, H. Natural strategies for photosynthetic light harvesting. Nature Chem. Biol. 10, 492–501 (2014)
Umena, Y., Kawakami, K., Shen, J.-R. & Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011)
Suga, M. et al. Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 517, 99–103 (2015)
Rhee, K.-H., Morris, E. P., Barber, J. & Kuhlbrandt, W. Three-dimensional structure of the plant photosystem II reaction centre at 8 Å resolution. Nature 396, 283–286 (1998)
Hankamer, B., Morris, E. P. & Barber, J. Revealing the structure of the oxygen-evolving core dimer of photosystem II by cryoelectron crystallography. Nature Struct. Mol. Biol. 6, 560–564 (1999)
Hankamer, B., Morris, E., Nield, J., Gerle, C. & Barber, J. Three-dimensional structure of the photosystem II core dimer of higher plants determined by electron microscopy. J. Struct. Biol. 135, 262–269 (2001)
Liu, Z. et al. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287–292 (2004)
Standfuss, J., Terwisscha van Scheltinga, A. C., Lamborghini, M. & Kühlbrandt, W. Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Å resolution. EMBO J. 24, 919–928 (2005)
Pan, X. et al. Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nature Struct. Mol. Biol. 18, 309–315 (2011)
Nield, J. & Barber, J. Refinement of the structural model for the Photosystem II supercomplex of higher plants. Biochim. Biophys. Acta 1757, 353–361 (2006)
Caffarri, S., Kouril, R., Kereiche, S., Boekema, E. J. & Croce, R. Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3063 (2009)
Drop, B. et al. Light-harvesting complex II (LHCII) and its supramolecular organization in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1837, 63–72 (2014)
Guskov, A. et al. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nature Struct. Mol. Biol. 16, 334–342 (2009)
Ferreira, K. N., Iverson, T. M., Maghlaoui, K., Barber, J. & Iwata, S. Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 (2004)
Garcia-Cerdán, J. G. et al. The PsbW protein stabilizes the supramolecular organization of photosystem II in higher plants. Plant J. 65, 368–381 (2011)
Ago, H. et al. Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J. Biol. Chem. 291, 5676–5687 (2016)
Shi, L. X. & Schroder, W. P. The low molecular mass subunits of the photosynthetic supracomplex, photosystem II. Biochim. Biophys. Acta 1608, 75–96 (2004)
Ballottari, M., Mozzo, M., Croce, R., Morosinotto, T. & Bassi, R. Occupancy and functional architecture of the pigment binding sites of photosystem II antenna complex Lhcb5. J. Biol. Chem. 284, 8103–8113 (2009)
Bricker, T. M., Roose, J. L., Fagerlund, R. D., Frankel, L. K. & Eaton-Rye, J. J. The extrinsic proteins of Photosystem II. Biochim. Biophys. Acta 1817, 121–142 (2012)
Ido, K. et al. Cross-linking evidence for multiple interactions of the PsbP and PsbQ proteins in a higher plant photosystem II supercomplex. J. Biol. Chem. 289, 20150–20157 (2014)
Mummadisetti, M. P. et al. Use of protein cross-linking and radiolytic footprinting to elucidate PsbP and PsbQ interactions within higher plant Photosystem II. Proc. Natl Acad. Sci. USA 111, 16178–16183 (2014)
Kapazoglou, A., Sagliocco, F. & Dure, L. PSII-T, a new nuclear encoded lumenal protein from photosystem II: targeting and processing in isolated chloroplasts. J. Biol. Chem. 270, 12197–12202 (1995)
Shabestari, M. H., Wolfs, C. J. A. M. & Spruijt, R. B. van Amerongen, H. & Huber, M. Exploring the structure of the 100 amino-acid residue long N-terminus of the plant antenna protein CP29. Biophys. J. 106, 1349–1358 (2014)
Qin, X., Suga, M., Kuang, T. & Shen, J. R. Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348, 989–995 (2015)
Mazor, Y., Borovikova, A. & Nelson, N. The structure of plant photosystem I super-complex at 2.8 Å resolution. eLife 4, e07433 (2015)
Caffarri, S., Passarini, F., Bassi, R. & Croce, R. A specific binding site for neoxanthin in the monomeric antenna proteins CP26 and CP29 of Photosystem II. FEBS Lett. 581, 4704–4710 (2007)
Yakushevska, A. E. et al. The structure of photosystem II in Arabidopsis: localization of the CP26 and CP29 antenna complexes. Biochemistry 42, 608–613 (2003)
de Bianchi, S. et al. Arabidopsis mutants deleted in the light-harvesting protein Lhcb4 have a disrupted photosystem II macrostructure and are defective in photoprotection. Plant Cell 23, 2659–2679 (2011)
Ruf, S., Biehler, K. & Bock, R. A small chloroplast-encoded protein as a novel architectural component of the light-harvesting antenna. J. Cell Biol. 149, 369–378 (2000)
Baena-González, E., Gray, J. C., Tyystjärvi, E., Aro, E.-M. & Mäenpää, P. Abnormal regulation of photosynthetic electron transport in a chloroplast ycf9 inactivation mutant. J. Biol. Chem. 276, 20795–20802 (2001)
Swiatek, M. et al. The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. Plant Cell 13, 1347–1368 (2001)
Remelli, R., Varotto, C., Sandona, D., Croce, R. & Bassi, R. Chlorophyll binding to monomeric light-harvesting complex. A mutation analysis of chromophore-binding residues. J. Biol. Chem. 274, 33510–33521 (1999)
Rogl, H. & Kühlbrandt, W. Mutant trimers of light-harvesting complex II exhibit altered pigment content and spectroscopic features. Biochemistry 38, 16214–16222 (1999)
Novoderezhkin, V. I., Palacios, M. A., van Amerongen, H. & van Grondelle, R. Excitation dynamics in the LHCII complex of higher plants: modeling based on the 2.72 Å crystal structure. J. Phys. Chem. B 109, 10493–10504 (2005)
Novoderezhkin, V., Marin, A. & van Grondelle, R. Intra- and inter-monomeric transfers in the light harvesting LHCII complex: the Redfield-Förster picture. Phys. Chem. Chem. Phys. 13, 17093–17103 (2011)
Sun, R. et al. Direct energy transfer from the major antenna to the photosystem II core complexes in the absence of minor antennae in liposomes. Biochim. Biophys. Acta 1847, 248–261 (2015)
Dall’Osto, L., Ünlü, C., Cazzaniga, S. & van Amerongen, H. Disturbed excitation energy transfer in Arabidopsis thaliana mutants lacking minor antenna complexes of photosystem II. Biochim. Biophys. Acta 1837, 1981–1988 (2014)
Mozzo, M., Passarini, F., Bassi, R., van Amerongen, H. & Croce, R. Photoprotection in higher plants: The putative quenching site is conserved in all outer light-harvesting complexes of Photosystem II. Biochim. Biophys. Acta 1777, 1263–1267 (2008)
Ahn, T. K. et al. Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320, 794–797 (2008)
Pascal, A. A. et al. Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature 436, 134–137 (2005)
Caffarri, S., Broess, K., Croce, R. & van Amerongen, H. Excitation energy transfer and trapping in higher plant Photosystem II complexes with different antenna sizes. Biophys. J. 100, 2094–2103 (2011)
Bennett, D. I. G., Amarnath, K. & Fleming, G. R. A. Structure-based model of energy transfer reveals the principles of light harvesting in photosystem II supercomplexes. J. Am. Chem. Soc. 135, 9164–9173 (2013)
Schägger, H. Tricine-SDS-PAGE. Nature Protocols 1, 16–22 (2006)
Färber, A., Young, A. J., Ruban, A. V., Horton, P. & Jahns, P. Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants (the relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching). Plant Physiol. 115, 1609–1618 (1997)
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods 10, 584–590 (2013)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Scheres, S. H. W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Allahverdiyeva, Y. et al. Insights into the function of PsbR protein in Arabidopsis thaliana. Biochim. Biophys. Acta 1767, 677–685 (2007)
Koua, F. H. M., Umena, Y., Kawakami, K. & Shen, J.-R. Structure of Sr-substituted photosystem II at 2.1 Å resolution and its implications in the mechanism of water oxidation. Proc. Natl Acad. Sci. USA 110, 3889–3894 (2013)
Li, X. P. et al. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395 (2000)
Fan, M. et al. Crystal structures of the PsbS protein essential for photoprotection in plants. Nature Struct. Mol. Biol. 22, 729–735 (2015)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Green, B. R. & Durnford, D. G. The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 685–714 (1996)
Das, S. K. & Frank, H. A. Pigment compositions, spectral properties, and energy transfer efficiencies between the xanthophylls and chlorophylls in the major and minor pigment−protein complexes of photosystem II. Biochemistry 41, 13087–13095 (2002)
Pascal, A. et al. Spectroscopic characterization of the spinach Lhcb4 protein (CP29), a minor light-harvesting complex of photosystem II. Eur. J. Biochem. 262, 817–823 (1999)
van Amerongen, H. et al. Spectroscopic characterization of CP26, a chlorophyll ab binding protein of the higher plant Photosystem II complex. Biochim. Biophys. Acta 1188, 227–234 (1994)
Acknowledgements
We thank J. P. Zhang and X. L. Zhao for their assistance in preparing thylakoid samples. Cryo-EM data collection was carried out at the Center for Biological Imaging, Core Facilities for Protein Science at the Institute of Biophysics (IBP), Chinese Academy of Sciences (CAS), and at the National Center for Protein Science Shanghai (NCPSS), Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences/Shanghai Science Research Center, Chinese Academy of Sciences, Shanghai, China. We thank X. J. Huang, G. Ji, W. Ding, F. Sun, and other staff members at the Center for Biological Imaging (IBP, CAS); L. L. Kong, X. Y. Shi, Y. N. He, J. P. Ding, and M. Lei for their support during data collection; X. B. Liang and X. M. An for support in organizing data collection trips; F. L. Zhang, J. Zhou, and Y. Li for support in measuring the oxygen evolution activity; L. L. Niu and X. Ding for mass spectrometry; J. H. Li for assistance in fluorescence measurement; R. Bassi, A. Pinnola and R. Croce for sharing experiences in purifying plant PSII–LHCII supercomplexes; and Y. Xiang for advice on cryo-EM sample preparation and structure refinement. The project was funded by National 973 project grant 2011CBA00900, the Strategic Priority Research Program of CAS (XDB08020302) and National Natural Science Foundation of China (31570724, 31270793 and 31170703). Z.L. and X.Z. received scholarships from the ‘National Thousand (Young) Talents Program’ from the Office of Global Experts Recruitment in China.
Author information
Authors and Affiliations
Contributions
X.W., X.S., P.C. and X.L. purified the spinach PSII–LHCII supercomplex; M.L. and P.C. characterized the spectroscopic features, protein and pigment contents, and oxygen-evolving activity of the samples; X.W., X.S. and X.Z. collected and processed cryo-EM data; X.Z. reconstructed the 3.2 Å resolution map and supervised cryo-EM structure determination; X.W. and Z.L. built and refined the structure model; X.W., M.L., X.Z. and Z.L. analysed the structure; Z.L. and W.C. conceived and coordinated the project; and the manuscript was written by X.W., M.L., X.Z. and Z.L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks Roberta Croce, Jian-Ren Shen and Thomas Walz for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Purification and characterization of the spinach PSII–LHCII supercomplex.
a, Sucrose gradient of solubilized grana membranes. The membrane preparations were first washed with 1 mM (left) or 5 mM (right) EDTA before being solubilized by α-DDM for further purification through sucrose-gradient ultracentrifugation. The content of each band is indicated based on the absorption spectrum and SDS–PAGE results, and by comparing to previously published data. The B9 fraction obtained from the grana membrane washed with 1 mM EDTA was used for cryo-EM. Note that the grana membranes washed with 1 mM EDTA yielded less B5, B6 and B7 than the sample treated with 5 mM EDTA. b, SDS–PAGE analysis of the sucrose gradient fractions. The protein composition of each Coomassie band was indicated based on the mass spectrometry and proteomics data analysis. For gel source data, see Supplementary Fig. 1. c, Room-temperature absorption spectrum of B9 sample used for cryo-EM. Its spectrum (B9 1 mM) is compared to those of B7 (dimeric PSII core without LHCII attached; B7 5 mM) and B9 samples (B9 5 mM) fractionated from grana washed with 5 mM EDTA. Note that B9 from grana membranes washed with 1 mM EDTA showed higher peaks at 470 and 650 nm, indicating that this fraction contains higher Chl b content (from LHCIIs) than the other two. The spectra are normalized to the maximum in the red region. d, Fluorescence emission spectra of B9 sample measured at room temperature. The maximum emissions were at 681 nm (upon excitation of Chl a at 436 nm), 680 nm (upon excitation of Chl b at 473 nm) and 681 nm (upon excitation of carotenoids at 500 nm). Overlapping of these three spectra suggests that nearly all pigments in the B9 sample are well coupled and no free pigments are present. e, Pigment content analysis of B9 sample by HPLC. Based on the characteristic absorption spectrum of each peak fraction, the six major pigment peaks separated from the B9 sample are identified as neoxanthin (Neo), violaxanthin (Vio), lutein (Lut), Chl b, Chl a and β-carotene (β-car).
Extended Data Figure 2 Evaluation of the resolution of the cryo-EM structure of the spinach PSII–LHCII supercomplex.
a, Fourier shell correlation (FSC) plots. Blue, gold-standard FSC curve with a value of 0.143 at 3.2 Å resolution; red, FSC curve calculated between the cryo-EM map and the refined structure model of the PSII–LHCII supercomplex. The map-model FSC has a value of 0.5 at 3.3 Å resolution. b, Local resolutions of the cryo-EM map of the spinach PSII–LHCII supercomplex estimated by Resmap. Top, side view along the membrane plane with the luminal domain facing upwards. Bottom, bottom view from the luminal side and approximately along the membrane normal (or C2 axis). c, The statistics of the structural model of the spinach PSII–LHCII supercomplex refined against the 3.2 Å resolution cryo-EM map.
Extended Data Figure 3 Structures of the large and small intrinsic subunits of spinach PSII.
a, The four large intrinsic subunits of the spinach PSII core superposed on the corresponding subunits of the TvPSII core. The protein backbones and cofactors are shown as ribbon and stick models, respectively. Silver, spinach PSII core subunits; green, TvPSII core subunits. b, The locations of 12 low-molecular-mass intrinsic subunits in the spinach PSII–LHCII supercomplex. These subunits are coloured and the rest of the supercomplex is grey. c, The densities for the low-molecular-mass intrinsic subunits are shown as blue meshes. The corresponding models are shown as cyan sticks.
Extended Data Figure 4 Cryo-EM densities and structures of the extrinsic subunits.
a, Cryo-EM densities of PsbO, PsbP, PsbQ and PsbTn. b, The binding sites of spinach PsbP, PsbQ and PsbO compared to those of the extrinsic subunits in CcPSII and TvPSII. The spinach PSII core is shown at an angle identical to that of the CcPSII/TvPSII core. PDB codes: 4YUU (CcPSII); 3WU2 (TvPSII). c, Superposition of PsbP bound in the supercomplex with the isolated PsbP. Colour code: yellow, PsbP in the supercomplex; blue, isolated PsbP (PDB code: 4RTI). Loop 3A and Loop 4A indicate the loop regions between Lys90 and Ala111 and between Arg134 and Gly142, respectively. Note the conformational change in Loop 3A (arrow) when PsbP binds to the PSII core. d, Structure of PsbQ bound in the supercomplex superposed with the isolated PsbQ. Green, PsbQ in the supercomplex; red, isolated PsbQ (PDB code: 1VYK). Note the conformational change in the elongated N-terminal region from a folded state to an extended form (arrow) when PsbQ binds to the PSII core.
Extended Data Figure 5 Cryo-EM density and structure of LHCII.
a, Cryo-EM densities of the LHCII trimer in the supercomplex. Stereo pairs are shown and the view is along the membrane plane. b, Superposition of the cryo-EM structure of an LHCII monomer with the previous crystal structure (PDB code: 1RWT). The protein backbone is shown as ribbon diagrams and the cofactors are displayed as stick models. Green, cryo-EM structure; yellow, crystal structure.
Extended Data Figure 6 Cryo-EM density and structure of CP29.
a, Stereo image of the cryo-EM density of CP29 bound in the PSII–LHCII supercomplex. b, Superposition of cryo-EM structure of full-length CP29 with the previous crystal structure. Note: Chls a601 and a616 are newly observed in the cryo-EM structure of CP29. Chl a601 might account for the electron density of Chl a615 observed in the crystal structure of spinach CP29 (ref. 14). Compared to Chl a601, a615 is much closer to a611 owing to the loss of the N-terminal domain caused by proteolysis. Chl b614 is a peripheral chlorophyll found in the crystal structure, but is probably lost during purification and therefore not observed in the cryo-EM structure. Orange, cryo-EM structure; cyan, crystal structure. c, Superposition of CP29 (orange) with the structure of an LHCII monomer (green). For the cofactors, only Chl a601 is shown; the others are omitted for clarity. d, Cryo-EM density of Chl a601 in CP29. e, Cryo-EM density of Chl a616 at the interface between CP29 and CP47. f, Superposition of Lhca3 and Lhca4 structures with that of CP29 in the PSII–LHCII supercomplex. Chl a616 (CP29) and a617 (Lhca3/4) molecules are shown as stick models; the other cofactors are omitted for clarity. Orange, CP29; magenta, Lhca3; blue, Lhca4. PDB codes: 4XK8, Lhca3 and Lhca4 from the PSI–LHCI supercomplex; 1RWT, LHCII.
Extended Data Figure 7 Cryo-EM density and structure of CP26 bound in the PSII–LHCII supercomplex.
a, Stereo images of the density and overall structure of CP26. The density is shown as grey meshes and the model is in purple. The protein backbone is shown as a ribbon model; the cofactors are presented as stick models. b, The densities for Chl b601, Chl a604, Chl b607 and Chl b608 in CP26. These four chlorophylls were not predicted in the previous work, but are clearly present in the structure. c, The density for three carotenoids in CP26. Note that the density for the epoxidized head group of neoxanthin is clearly visible, while the rest of it is fairly weak (presumably owing to low occupancy or high flexibility).
Extended Data Figure 8 Cryo-EM densities of various cofactors bound in the spinach PSII–LHCII supercomplex.
a, The densities of chlorophylls, carotenoids, Mn4CaO5 and plastoquinone molecules. b, The potential lipid densities at the interfacial regions between adjacent antenna complexes. The interfaces between LHCII and PsbW, CP26 and CP43, and LHCII and CP29 are shown from left to right. Red arrows indicate the positions of potential lipid densities. The cryo-EM densities are displayed as grey meshes and the atomic models for interpretation of the densities are shown as sticks and bullets.
Supplementary information
Supplementary Figure 1
This file contains the source data for Extended Data Figure 1b. The boxed area is cropped and displayed in the Extended Data Figure 1b. (PDF 140 kb)
Rights and permissions
About this article
Cite this article
Wei, X., Su, X., Cao, P. et al. Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution. Nature 534, 69–74 (2016). https://doi.org/10.1038/nature18020
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18020
This article is cited by
-
The nature of carotenoid S* state and its role in the nonphotochemical quenching of plants
Nature Communications (2024)
-
Azobenzene-based ultrathin peptoid nanoribbons for the potential on highly efficient artificial light-harvesting
Science China Chemistry (2024)
-
Uphill energy transfer mechanism for photosynthesis in an Antarctic alga
Nature Communications (2023)
-
Generation and physiological characterization of genome-edited Nicotiana benthamiana plants containing zeaxanthin as the only leaf xanthophyll
Planta (2023)
-
Role of hydrogen-bond networks on the donor side of photosynthetic reaction centers from purple bacteria
Biophysical Reviews (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.