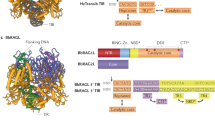
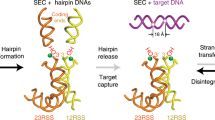
Abstract
V(D)J recombination in the vertebrate immune system generates a highly diverse population of immunoglobulins and T-cell receptors by combinatorial joining of segments of coding DNA. The RAG1–RAG2 protein complex initiates this site-specific recombination by cutting DNA at specific sites flanking the coding segments. Here we report the crystal structure of the mouse RAG1–RAG2 complex at 3.2 Å resolution. The 230-kilodalton RAG1–RAG2 heterotetramer is ‘Y-shaped’, with the amino-terminal domains of the two RAG1 chains forming an intertwined stalk. Each RAG1–RAG2 heterodimer composes one arm of the ‘Y’, with the active site in the middle and RAG2 at its tip. The RAG1–RAG2 structure rationalizes more than 60 mutations identified in immunodeficient patients, as well as a large body of genetic and biochemical data. The architectural similarity between RAG1 and the hairpin-forming transposases Hermes and Tn5 suggests the evolutionary conservation of these DNA rearrangements.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sakano, H., Huppi, K., Heinrich, G. & Tonegawa, S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280, 288–294 (1979)
Lewis, S. M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv. Immunol. 56, 27–150 (1994)
Gellert, M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71, 101–132 (2002)
Schatz, D. G. & Swanson, P. C. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45, 167–202 (2011)
Deriano, L. & Roth, D. B. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47, 433–455 (2013)
Boboila, C., Alt, F. W. & Schwer, B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv. Immunol. 116, 1–49 (2012)
Oettinger, M. A., Schatz, D. G., Gorka, C. & Baltimore, D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248, 1517–1523 (1990)
Schatz, D. G., Oettinger, M. A. & Baltimore, D. The V(D)J recombination activating gene, RAG-1. Cell 59, 1035–1048 (1989)
McBlane, J. F. et al. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83, 387–395 (1995)
Agrawal, A., Eastman, Q. M. & Schatz, D. G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394, 744–751 (1998)
Hiom, K., Melek, M. & Gellert, M. DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell 94, 463–470 (1998)
Lee, Y. N. et al. A systematic analysis of recombination activity and genotype-phenotype correlation in human recombination-activating gene 1 deficiency. J. Allergy Clin. Immunol. 133, 1099–1108 (2014)
Piirilä, H., Valiaho, J. & Vihinen, M. Immunodeficiency mutation databases (IDbases). Hum. Mutat. 27, 1200–1208 (2006)
Cuomo, C. A. & Oettinger, M. A. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 22, 1810–1814 (1994)
Sadofsky, M. J., Hesse, J. E. & Gellert, M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 22, 1805–1809 (1994)
Sadofsky, M. J., Hesse, J. E., McBlane, J. F. & Gellert, M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 21, 5644–5650 (1993)
Silver, D. P., Spanopoulou, E., Mulligan, R. C. & Baltimore, D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl Acad. Sci. USA 90, 6100–6104 (1993)
Grundy, G. J. et al. Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex. Mol. Cell 35, 217–227 (2009)
Jones, J. M. & Gellert, M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl Acad. Sci. USA 98, 12926–12931 (2001)
Liu, Q., Zhang, Z. & Hendrickson, W. A. Multi-crystal anomalous diffraction for low-resolution macromolecular phasing. Acta Crystallogr. D 67, 45–59 (2011)
Gigi, V. et al. RAG2 mutants alter DSB repair pathway choice in vivo and illuminate the nature of ‘alternative NHEJ’. Nucleic Acids Res. 42, 6352–6364 (2014)
Yin, F. F. et al. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nature Struct. Mol. Biol. 16, 499–508 (2009)
Fugmann, S. D., Villey, I. J., Ptaszek, L. M. & Schatz, D. G. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell 5, 97–107 (2000)
Kim, D. R., Dai, Y., Mundy, C. L., Yang, W. & Oettinger, M. A. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 13, 3070–3080 (1999)
Landree, M. A., Wibbenmeyer, J. A. & Roth, D. B. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 13, 3059–3069 (1999)
Nesmelova, I. V. & Hackett, P. B. DDE transposases: Structural similarity and diversity. Adv. Drug Deliv. Rev. 62, 1187–1195 (2010)
Swanson, P. C. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21, 449–458 (2001)
Yang, W. & Steitz, T. A. Recombining the structures of HIV integrase, RuvC and RNase H. Structure 3, 131–134 (1995)
Gwyn, L. M., Peak, M. M., De, P., Rahman, N. S. & Rodgers, K. K. A zinc site in the C-terminal domain of RAG1 is essential for DNA cleavage activity. J. Mol. Biol. 390, 863–878 (2009)
Rodgers, K. K. et al. A zinc-binding domain involved in the dimerization of RAG1. J. Mol. Biol. 260, 70–84 (1996)
Aravind, L. & Koonin, E. V. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287, 1023–1040 (1999)
Callebaut, I. & Mornon, J. P. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell. Mol. Life Sci. 54, 880–891 (1998)
Swanson, P. C. & Desiderio, S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 19, 3674–3683 (1999)
Huye, L. E., Purugganan, M. M., Jiang, M. M. & Roth, D. B. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell. Biol. 22, 3460–3473 (2002)
Ko, J. E., Kim, C. W. & Kim, D. R. Amino acid residues in RAG1 responsible for the interaction with RAG2 during the V(D)J recombination process. J. Biol. Chem. 279, 7715–7720 (2004)
Hare, S., Gupta, S. S., Valkov, E., Engelman, A. & Cherepanov, P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464, 232–236 (2010)
Hickman, A. B. et al. Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell 158, 353–367 (2014)
Montaño, S. P., Pigli, Y. Z. & Rice, P. A. The mu transpososome structure sheds light on DDE recombinase evolution. Nature 491, 413–417 (2012)
Richardson, J. M., Colloms, S. D., Finnegan, D. J. & Walkinshaw, M. D. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell 138, 1096–1108 (2009)
Steiniger-White, M., Rayment, I. & Reznikoff, W. S. Structure/function insights into Tn5 transposition. Curr. Opin. Struct. Biol. 14, 50–57 (2004)
Grundy, G. J., Hesse, J. E. & Gellert, M. Requirements for DNA hairpin formation by RAG1/2. Proc. Natl Acad. Sci. USA 104, 3078–3083 (2007)
Lu, C. P., Sandoval, H., Brandt, V. L., Rice, P. A. & Roth, D. B. Amino acid residues in Rag1 crucial for DNA hairpin formation. Nature Struct. Mol. Biol. 13, 1010–1015 (2006)
Aidinis, V. et al. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1–RAG2. Mol. Cell. Biol. 19, 6532–6542 (1999)
van Gent, D. C., Hiom, K., Paull, T. T. & Gellert, M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 16, 2665–2670 (1997)
Kim, D. R. & Oettinger, M. A. Functional analysis of coordinated cleavage in V(D)J recombination. Mol. Cell. Biol. 18, 4679–4688 (1998)
Aricescu, A. R., Lu, W. & Jones, E. Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D 62, 1243–1250 (2006)
Swanson, P. C. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 22, 7790–7801 (2002)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Chenna, R. et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 (2003)
Schumacher, F. R., Sorrell, F. J., Alessi, D. R., Bullock, A. N. & Kurz, T. Structural and biochemical characterization of the KLHL3-WNK kinase interaction important in blood pressure regulation. Biochem. J. 460, 237–246 (2014)
Corbett, K. D., Shultzaberger, R. K. & Berger, J. M. The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc. Natl Acad. Sci. USA 101, 7293–7298 (2004)
Schuetz, C. et al. Lesson from hypomorphic recombination-activating gene (RAG) mutations: Why asymptomatic siblings should also be tested. J. Allergy Clin. Immunol. 133, 1211–1215 (2014)
Corneo, B. et al. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T-B-severe combined immune deficiency or Omenn syndrome. Blood 97, 2772–2776 (2001)
Dhingra, N. et al. Severe combined immunodeficiency caused by a new homozygous RAG1 mutation with progressive encephalopathy. Hematol. Oncol. Stem Cell Ther. 7, 44–49 (2014)
Acknowledgements
We thank G. Grundy and S. Ramon-Maiques for the pioneering work that made this study possible, and D. Leahy for editing the manuscript. The research was supported by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Author information
Authors and Affiliations
Contributions
M.-S.K. prepared SEC complexes and carried out crystallography. M.L. developed protocols for protein expression and initial crystallization. W.Y. and M.G. designed the project, and M.-S.K., W.Y. and M.G. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Structure determination.
a, The plot of correlation coefficient (CC) of anomalous signal versus resolution. The red line indicates the cutoff of CC = 0.3. Merging data from the two best crystals produced a better CC than merging data from all six crystals. The data processing procedure is outlined above the plot20. b, The SAD experimental map contoured at 1.3σ showed the content of an asymmetric unit. The Se anomalous map is contoured at 3.0σ in red. c, A typical crystal of RAG1–RAG2. d, The content of crystals was examined by protein and DNA denaturing gels after a thorough wash of the crystals and stained by Coomassie blue and SYBR green. To confirm the 1:1 molar ratio of 12 and 23RSS DNA, 32P-labelled input RSS DNAs and those in SEC complexes before and after crystallization are shown beneath the SYBR-green-stained DNA gel. e, Transposition assay of the purified SEC (RAG1–RAG2–12/23RSS DNA complex) used for crystallization. Supercoiled pUC19 (sc, with a small amount of open circle, oc) was the target; it was linearized by HindIII as a control. The SEC (0.25, 0.5 and 1.0 μM) was active in concerted transposition and thus linearizing pUC19. In contrast, RAG1–RAG2 or HMGB1 (0.5 µM) each alone was not active. f, Crystal packing of neighbouring RAG1–RAG2 complexes (shown in dark and light colours) occludes one nonamer-binding site in each heterotetramer of RAG1–RAG2.
Extended Data Figure 2 RAG2 core fragment (1–351 amino acids) is active.
a, Sequence alignment of RAG2 from mouse (320–387 amino acids), human, rat and Xenopus with predicted secondary structures shown above. b, Core RAG2 (1–387) and two further truncated RAG2 variants (1–351 and 1–367) were constructed with a non-cleavable N-terminal MBP tag and co-expressed with the tag-less core RAG1. The Coomassie blue R-250 stained SDS gel shows the purified RAG1–RAG2 complexes. c, Purified RAG1–RAG2 complexes with truncated RAG2 variants are equally active in cleaving a 32P-labelled 12RSS DNA (in the presence of a 23RSS and Mg2+, as examined by TBE-Urea gel). d, Elution profiles of RAG1–RAG2 (both long and short forms) complexed with DNA from Superdex-200 (S200) in a low salt buffer (50 mM HEPES pH 7.0, 60 mM KCl, 1 mM maltose and 2 mM DTT). Regardless of the length of RAG2, the major S200 eluant peak came out at the same time point and contained RAG1, RAG2 (1–351 or 1–387) and HMGB1 proteins, as shown in the SDS gel (right insert), as well as 12 and 23RSS oligonucleotides, as confirmed by a TBE-Urea gel stained by SYBR green (left insert).
Extended Data Figure 3 Comparison of RAG2 with β-propeller and β-pinwheel structures.
KLHL2 (PDB 4CHB)56 is selected to represent the β-propeller proteins, and the C-terminal domain (CTD) of GyrA (PDB 1SUU)57 is selected to represent the β-pinwheel structures. After superposition, RAG2 (a), KLHL2 (b) and GyrA (c) are shown side-by-side individually in two orthogonal views. Each structure is coloured from N to C terminus in blue to red rainbow colours. The loops in RAG2 that interact with RAG1 are labelled. The six β-blades are named by Roman numerals, I–VI, from N to C terminus; four β-strands in each blade are named by Arabic numerals, 1–4.
Extended Data Figure 4 Comparison of RAG1 and NBD–DNA complex.
a, The NBD in the RAG1–RAG2 core complex (blue and green) superimposes well with the published structure of the NBD–DNA complex (PDB 3GNA, protein coloured yellow)22. b, The twelve SCID/Omenn syndrome mutations in the NBD domain are mapped onto the crystal structure of the NBD–DNA complex. Six SCID/Omenn syndrome (R391 to R407) mutations are located on a positively charged surface patch that interacts with the nonamer; five remaining SCID/Omenn syndrome mutations (L408 to A441) appear to affect the structural integrity of the NBD, and R446 may interact with the spacer DNA in each RSS.
Extended Data Figure 5 Transposases that form a hairpin intermediate.
a–c, Hermes (PDB 4D1Q)37 (a), bacterial Tn5 (PDB code 1MUS)40 (b), and RAG1 dimers (c) are shown as ribbon diagrams in two orthogonal views, with the dyad perpendicular to the viewing plane (left) or in the plane (right). Each dimer consists of a cyan and a green subunit. The catalytic RNH domains are highlighted in pink, and the conserved catalytic residues are shown as red ball-and-sticks. The catalytic divalent metal ions are shown as green spheres if present. The DNAs, coloured in yellow (cleaved by the cyan subunit) and orange (cleaved by the green subunit), have similar orientations in the Hermes and Tn5 complexes (as indicated by the arrows). Arrows with dashed outlines indicate that the DNAs are in the back of the viewing plane. Notably, the pair of RNH domains is oriented similarly in all three cases. The predicted orientations of DNAs bound to RAG1 are indicated by the yellow and orange arrows, and the α-helices connected to the third catalytic carboxylates (shown in light purple) probably bridge two DNAs in RAG1 recombinase as in Hermes and Tn5.
Extended Data Figure 6 Transposases that do not form a hairpin intermediate.
a–c, Retroviral integrase from Prototype foamy virus (Pfv, PDB 3OS0)36 (a), bacterial MuA transposase (PDB code 4FCY)38 (b) and eukaryotic Mos1 mariner transposase (PDB 3HOT)39 (c) are shown in comparable views and same representations as Hermes, Tn5 and RAG1–RAG2 in Extended Data Fig. 5. Each catalytic dimer consists of a cyan and a green subunit. Two accessory subunits in Pfv are shown in light blue and green, and two accessory subunits of the MuA structure are omitted for clarity. The catalytic RNH domains are highlighted in pink. The DNAs, coloured in yellow (cleaved by the cyan subunit) and orange (cleaved by the green subunit), have similar orientations (within 30°) as indicated by the arrowheads, but each differs more than 90° from the corresponding DNA in Hermes or Tn5 transposase. The grey DNA in the MuA complex represents the target of transposition. Among these three recombinases, the α-helix that follows the third catalytic carboxylate (coloured in light purple) does not cross over to interact with a second DNA.
Extended Data Figure 7 Surface potential and conservation of RAG1–RAG2 complex.
a, Orthogonal views of the electrostatic potential surface of the RAG1–RAG2 structure. Blue indicates positive charges, and red negative. b, Orthogonal views of the molecular surface of RAG1–RAG2 with absolutely conserved residues highlighted in deep purple. The NBD is well conserved. The views with dyad in the plane here are related to the image shown in Fig. 5c by ∼50° rotation around the dyad.
Supplementary information
Animation of the SEC model
RAG1/2 is shown in surface representation. Blue and red represent positively and negatively charged surface potentials. The nonamers are modeled after the NBD-DNA complex structure (12bp, PDB: 3GNA), after domain superposition. Each 16bp DNA including the heptamer mimic is brought in by superposition of the Hermes-DNA complex (PDB: 4D1Q) with the RNH domain of each RAG1 subunit. The view shows the 12RSS-like DNA, where the 16bp and 12bp DNAs are nearly touching. The 23RSS-like DNA is in the back, and the two DNAs modeled are ~30Å apart. (MOV 4452 kb)
Rights and permissions
About this article
Cite this article
Kim, MS., Lapkouski, M., Yang, W. et al. Crystal structure of the V(D)J recombinase RAG1–RAG2. Nature 518, 507–511 (2015). https://doi.org/10.1038/nature14174
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14174
This article is cited by
-
Comparison of immunophenotypes between Rag2 knockout mice derived from two different sources
Laboratory Animal Research (2023)
-
A variant of RAG1 gene identified in severe combined immunodeficiency: a case report
BMC Pediatrics (2023)
-
The role of chromatin loop extrusion in antibody diversification
Nature Reviews Immunology (2022)
-
Structural insights into the evolution of the RAG recombinase
Nature Reviews Immunology (2022)
-
Dimers of DNA-PK create a stage for DNA double-strand break repair
Nature Structural & Molecular Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.