Abstract
Metazoans identify and eliminate bacterial pathogens in microbe-rich environments such as the intestinal lumen; however, the mechanisms are unclear. Host cells could potentially use intracellular surveillance or stress response programs to detect pathogens that target monitored cellular activities and then initiate innate immune responses1,2,3. Mitochondrial function is evaluated by monitoring mitochondrial protein import efficiency of the transcription factor ATFS-1, which mediates the mitochondrial unfolded protein response (UPRmt). During mitochondrial stress, mitochondrial import is impaired4, allowing ATFS-1 to traffic to the nucleus where it mediates a transcriptional response to re-establish mitochondrial homeostasis5. Here we examined the role of ATFS-1 in Caenorhabditis elegans during pathogen exposure, because during mitochondrial stress ATFS-1 induced not only mitochondrial protective genes but also innate immune genes that included a secreted lysozyme and anti-microbial peptides. Exposure to the pathogen Pseudomonas aeruginosa caused mitochondrial dysfunction and activation of the UPRmt. C. elegans lacking atfs-1 were susceptible to P. aeruginosa, whereas hyper-activation of ATFS-1 and the UPRmt improved clearance of P. aeruginosa from the intestine and prolonged C. elegans survival in a manner mainly independent of known innate immune pathways6,7. We propose that ATFS-1 import efficiency and the UPRmt is a means to detect pathogens that target mitochondria and initiate a protective innate immune response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Melo, J. A. & Ruvkun, G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452–466 (2012)
McEwan, D. L., Kirienko, N. V. & Ausubel, F. M. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 11, 364–374 (2012)
Dunbar, T. L., Yan, Z., Balla, K. M., Smelkinson, M. G. & Troemel, E. R. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 11, 375–386 (2012)
Wright, G., Terada, K., Yano, M., Sergeev, I. & Mori, M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp. Cell Res. 263, 107–117 (2001)
Nargund, A. M., Pellegrino, M. W., Fiorese, C. J., Baker, B. M. & Haynes, C. M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590 (2012)
Kim, D. H. et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626 (2002)
Twumasi-Boateng, K. et al. An age-dependent reversal in the protective capacities of JNK signaling shortens Caenorhabditis elegans lifespan. Aging Cell 11, 659–667 (2012)
Guarner, F. & Malagelada, J. R. Gut flora in health and disease. Lancet 361, 512–519 (2003)
Newton, K. & Dixit, V. M. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 4, http://dx.doi.org/10.1101/cshperspect.a006049 (2012)
Troemel, E. R. et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2, e183 (2006)
Kato, Y. et al. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J. 361, 221–230 (2002)
Nandakumar, M. & Tan, M. W. Gamma-linolenic and stearidonic acids are required for basal immunity in Caenorhabditis elegans through their effects on p38 MAP kinase activity. PLoS Genet. 4, e1000273 (2008)
Nicholas, H. R. & Hodgkin, J. Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol. Immunol. 41, 479–493 (2004)
De Smet, K. & Contreras, R. Human antimicrobial peptides: defensins, cathelicidins and histatins. Biotechnol. Lett. 27, 1337–1347 (2005)
O’Malley, Y. Q. et al. Subcellular localization of Pseudomonas pyocyanin cytotoxicity in human lung epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 284, L420–L430 (2003)
Gallagher, L. A. & Manoil, C. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183, 6207–6214 (2001)
Estes, K. A., Dunbar, T. L., Powell, J. R., Ausubel, F. M. & Troemel, E. R. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 107, 2153–2158 (2010)
Pukkila-Worley, R. et al. Stimulation of host immune defenses by a small molecule protects C. elegans from bacterial infection. PLoS Genet. 8, e1002733 (2012)
Tan, M. W., Mahajan-Miklos, S. & Ausubel, F. M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl Acad. Sci. USA 96, 715–720 (1999)
Gomes, L. C., Di Benedetto, G. & Scorrano, L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nature Cell Biol. 13, 589–598 (2011)
Feng, J., Bussiere, F. & Hekimi, S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644 (2001)
Kirienko, N. V. et al. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 13, 406–416 (2013)
Tan, M. W., Rahme, L. G., Sternberg, J. A., Tompkins, R. G. & Ausubel, F. M. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl Acad. Sci. USA 96, 2408–2413 (1999)
Richardson, C. E., Kooistra, T. & Kim, D. H. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463, 1092–1095 (2010)
Baker, B. M., Nargund, A. M., Sun, T. & Haynes, C. M. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 8, e1002760 (2012)
Walter, L., Baruah, A., Chang, H. W., Pace, H. M. & Lee, S. S. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS Biol. 9, e1001084 (2011)
Rauthan, M., Ranji, P., Aguilera Pradenas, N., Pitot, C. & Pilon, M. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proc. Natl Acad. Sci. USA 110, 5981–5986 (2013)
Kim, D. H. et al. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc. Natl Acad. Sci. USA 101, 10990–10994 (2004)
Runkel, E. D., Liu, S., Baumeister, R. & Schulze, E. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet. 9, e1003346 (2013)
Liu, Y., Samuel, B. S., Breen, P. C. & Ruvkun, G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508, 406–410 (2014)
Yoneda, T. et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 117, 4055–4066 (2004)
Rual, J. F. et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168 (2004)
Ehses, S. et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036 (2009)
Zhao, Q. et al. A mitochondrial specific stress response in mammalian cells. EMBO J. 21, 4411–4419 (2002)
Tan, M. W., Mahajan-Miklos, S. & Ausubel, F. M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl Acad. Sci. USA 96, 715–720 (1999)
Liu, Y., Samuel, B. S., Breen, P. C. & Ruvkun, G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508, 406–410 (2014)
Nargund, A. M., Pellegrino, M. W., Fiorese, C. J., Baker, B. M. & Haynes, C. M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590 (2012)
Kennedy, S., Wang, D. & Ruvkun, G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427, 645–649 (2004)
Benedetti, C., Haynes, C. M., Yang, Y., Harding, H. P. & Ron, D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 174, 229–239 (2006)
Kirienko, N. V. et al. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 13, 406–416 (2013)
Acknowledgements
We thank E. Troemel, M. Pilon, S. Mitani of the National Bioresource Project, and the Caenorhabditis Genetics Center for providing C. elegans strains. We thank D. Kim and J. Xavier for providing P. aeruginosa strains, and T. Langer and S. Troeder for mammalian reagents. This work was supported by the Ellison Medical Foundation, the Lucille Castori Center for Microbes, Inflammation and Cancer at MSKCC and the National Institutes of Health (R01AG040061) to C.M.H., (F32AI100501) to N.V.K. and (R01AI085581) to F. Ausubel.
Author information
Authors and Affiliations
Contributions
M.W.P., A.M.N., N.V.K., R.G. and C.J.F. performed experiments. M.W.P. and C.M.H. conceived of and planned the experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
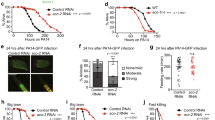
Extended Data Figure 1 Nuclear accumulation of ATFS-1 is required for UPRmt activation during P. aeruginosa exposure.
a, Representative photomicrographs of F35E12.5pr::gfp transgenic worms raised on control or spg-7(RNAi). No detectable increase in expression was observed following spg-7(RNAi) treatment. In contrast, strong expression of F35E12.5pr::gfp was observed following exposure to P. aeruginosa compared to E. coli controls. Scale bar, 0.5 mm. b, Wild-type or atfs-1(tm4525);hsp-60pr::gfp worms on E. coli or P. aeruginosa. Lower panels are magnified views of the intestine showing enhanced expression of hsp-60pr::gfp (asterisks). Scale bars, 0.05 mm. c, Diagrams of wild-type ATFS-1 (ATFS-1FL) and ATFS-1 with a mutated nuclear localization signal (ATFS-1ΔNLS). d, Photomicrographs of atfs-1(tm4525);hsp-60pr::gfp worms expressing ATFS-1FL or ATFS-1ΔNLS via the hsp-16 promoter exposed to E. coli or P. aeruginosa. Scale bar, 0.1 mm.
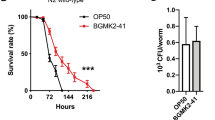
Extended Data Figure 2 Multiple P. aeruginosa virulence genes contribute to UPRmt activation.
a, b, Expression of hsp-60 and hsp-6 mRNA for glp-4(bn2) worms exposed to E. coli or P. aeruginosa liquid-killing using qRT–PCR (n = 3, ± s.d.). Fold inductions are normalized to wild-type E. coli test group, *P < 0.05 (Student’s t-test). c, Quantification of survival for glp-4(bn2) worms raised on control or atfs-1(RNAi) and exposed to P. aeruginosa liquid-killing, *P < 0.0001 (Student’s t-test). d, List of P. aeruginosa toxin mutants. e, Quantification of the proportion of worms showing increased hsp-6pr::gfp expression in the intestine under slow-killing conditions. Exposure to P. aeruginosa caused hsp-6pr::gfp induction (n = 3, ± s.e.m.), *P < 0.05 (Student’s t-test). However, exposure to P. aeruginosa with mutations in the pvdA, pvdD, pvdF, phzM, hcnB, or hcnC toxin genes resulted in relatively less UPRmt activation (n = 3, ± s.e.m.), **P < 0.05 (Student t-test).
Extended Data Figure 3 Intestinal accumulation of lys-2 during mitochondrial stress and P. aeruginosa exposure requires ATFS-1.
a, Representative photomicrographs of wild-type and atfs-1(tm4919) worms carrying the lys-2pr::gfp transgene raised on control or spg-7(RNAi). Scale bar, 0.1 mm. b, Representative photomicrographs of wild-type and atfs-1(tm4919) worms carrying the lys-2pr::gfp transgene exposed to E. coli or P. aeruginosa. Scale bar, 0.1 mm.
Extended Data Figure 4 ATFS-1 partially regulates zip-2 expression during P. aeruginosa exposure.
a, Expression levels of zip-2 mRNA in wild-type or atfs-1(tm4525) worms raised on E. coli or P. aeruginosa using qRT–PCR (n = 3, ± s.d.), * P < 0.05 (Student’s t test). b, Schematic diagram of the atfs-1 genomic open reading frame showing positions of exons 1–8 (boxes) and locations of the tm4525 (ref. 5) and tm4919 deletions in red. The tm4919 allele is a 334 base pair deletion beginning 107 base pairs upstream of the atfs-1 start codon and ends within the second intron of the atfs-1 genomic open reading frame. c, Representative photomicrographs of a germline in wild-type and atfs-1(tm4919) worms. Scale bar, 0.02 mm.
Extended Data Figure 5 ATFS-1 is not required for pathogen avoidance during P. aeruginosa exposure.
a, Quantification of avoidance behaviour for wild-type and atfs-1(tm4919) worms raised on E. coli or P. aeruginosa, expressed as a percentage of the number of animals off the bacterial lawn relative to the total number of worms (n = 4, ± s.d.). *P < 0.0001, **P = 0.1914 (Student’s t-test). b, Quantification of avoidance behaviour for wild-type worms raised on control or spg-7(RNAi) and exposed to E. coli or P. aeruginosa, expressed as a percentage of the number of animals off the bacterial lawn relative to the total number of worms (n = 3, ± s.d.). *P < 0.0001, **P = 0.8706 (Student’s t-test). c, Representative photomicrographs illustrating the scored level of infection for P. aeruginosa colonization assay using P. aeruginosa–GFP. Three categories of P. aeruginosa–GFP infection were used: none/mild, moderate and strong. Scale bar, 0.1 mm. d, Representative photomicrographs of wild-type and atfs-1(tm4919) worms raised on spg-7(RNAi) and exposed to a lawn of P. aeruginosa–GFP that completely covered the surface of the slow-killing plate for 24 h. Images are overlays of DIC and GFP. Scale bar, 0.1 mm. e, Quantification of P. aeruginosa intestinal colonization as shown in Extended Data Fig. 5d. White, grey and black bars denote no/mild infection, moderate infection and strong infection, respectively. Forty worms were analysed per treatment. f, Survival analysis of glp-4(bn2) and atfs-1(tm4919); glp-4(bn2) worms raised on control or spg-7(RNAi) and exposed to P. aeruginosa. Statistics for each survival analysis are presented in Extended Data Table 3. g, Quantification of pharyngeal pumping rate per minute for wild-type worms raised on control or spg-7(RNAi) (n = 10, + s.d.). n.s., no significant difference (P = 0.10; Student’s t-test).
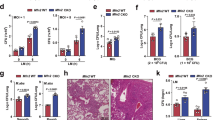
Extended Data Figure 6 atfs-1(et18) gain of function mutant worms induce innate immune gene expression in the absence of mitochondrial stress.
a–d, Expression levels of abf-2, lys-2, clec-4 and clec-65 mRNA in wild-type or atfs-1(et18) worms using qRT–PCR (n = 3, ± s.d.), *P < 0.05 (Student’s t test). e, Representative photomicrographs of wild-type and atfs-1(et18) worms carrying the irg-1pr::gfp transgene raised on control or zip-2(RNAi). Scale bar, 0.10 mm. f, Survival analysis of wild-type and atfs-1(et18) worms raised on control or lys-2(RNAi) and exposed to P. aeruginosa. Statistics for each survival analysis are presented in Extended Data Table 3.
Extended Data Figure 7 Mitochondrial protective and innate immune gene induction contributes to ATFS-1-mediated resistance to P. aeruginosa infection.
a, Representative photomicrographs of wild-type hsp-60pr::gfp worms raised on control, atp-2(RNAi), spg-7(RNAi), eft-2(RNAi), sca-1(RNAi), T25B9.9(RNAi) or T08A11.2(RNAi). Scale bar is 0.1 mm. b, Representative photomicrographs of wild-type irg-1pr::gfp worms raised on control, atp-2(RNAi), eft-2(RNAi), sca-1(RNAi), T25B9.9(RNAi) or T08A11.2(RNAi). Scale bar is 0.1 mm. c, Survival analysis of wild-type worms raised on control, atp-2(RNAi), eft-2(RNAi), sca-1(RNAi), T25B9.9(RNAi) or T08A11.2(RNAi) and exposed to P. aeruginosa. Statistics for each survival analysis are presented in Extended Data Table 3. d, Survival analysis of wild-type worms raised on control, atp-2(RNAi), eft-2(RNAi), sca-1(RNAi), T25B9.9(RNAi) or T08A11.2(RNAi) and exposed to E. coli. Statistics for each survival analysis are presented in Extended Data Table 3. e, Representative photomicrographs of wild-type or kgb-1(km21);hsp-60pr::gfp worms raised on E. coli plates with or without 30 μg ml−1 ethidium bromide, suggesting that the KGB-1 Jun kinase pathway negatively regulates the UPRmt during mitochondrial stress29. Scale bar, 0.5 mm.
Rights and permissions
About this article
Cite this article
Pellegrino, M., Nargund, A., Kirienko, N. et al. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516, 414–417 (2014). https://doi.org/10.1038/nature13818
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13818
This article is cited by
-
Mitochondrial perturbation in immune cells enhances cell-mediated innate immunity in Drosophila
BMC Biology (2024)
-
Mitochondrial aconitase suppresses immunity by modulating oxaloacetate and the mitochondrial unfolded protein response
Nature Communications (2023)
-
ATFS-1 counteracts mitochondrial DNA damage by promoting repair over transcription
Nature Cell Biology (2023)
-
Lysosome-related organelles promote stress and immune responses in C. elegans
Communications Biology (2023)
-
The homeodomain transcription factor CEH-37 regulates PMK-1/p38 MAPK pathway to protect against intestinal infection via the phosphatase VHP-1
Cellular and Molecular Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.