Abstract
Caspases are a family of cysteine proteases widely known as the principal mediators of the apoptotic cell death response, but considerably less so as the contributors to the regulation of pathways outside cellular demise. In regards to autophagy, the modulatory roles of caspases have only recently begun to be adequately described. In contrast to apoptosis, autophagy promotes cell survival by providing energy and nutrients through the lysosomal degradation of cytoplasmic constituents. Under basal conditions autophagy and apoptosis cross-regulate each other through an elaborate network of interconnections which also includes the interplay between autophagy-related proteins (ATGs) and caspases. In this review we focus on the effects of this crosstalk at the cellular level, as we aim to concentrate the main observations from research conducted so far on the fine-tuning of autophagy by caspases. Several members of this protease-family have been found to directly interact with key ATGs involved in different tiers across the autophagic cascade. Therefore, we firstly outline the core mechanism of macroautophagy in brief. In an effort to emphasize the importance of the intricate cross-regulation of ATGs and caspases, we also present examples of autophagy’s contribution to apoptotic cell death during development.
Similar content being viewed by others
Facts
-
Caspases have been shown to directly interact with core autophagy proteins.
-
Autophagy is implicated in many physiological and pathological processes, where apoptosis is also involved.
Open Questions
-
Does control of autophagy by caspases represent a central or a supplementary mode of regulation of this pathway?
-
Is caspase activation required to control autophagy?
-
To what extent do caspases affect autophagy and how exactly their effects on the pathway vary across different contexts?
Apoptosis is the tightly controlled process for an orderly executed cell death, and its key mediators, the caspases (Cysteine-dependent aspartate-specific proteases), are known for their indispensable part in the cascade. They exist as inactive precursors within cells, activated by oligomerization, cleavage of regulatory pro-domains, and/or dissociation from endogenous inhibitors.1, 2, 3 In mammals, the ten major caspases (of the 14 characterized so far) are classified based on their primary apoptotic role, as initiators (caspase-2,-8,-9, and -10), or executioners (caspase-3, -6, and -7).1, 2, 3 The third group consists of inflammatory caspases (caspase-1, -4, and -5) which are involved in ‘pyroptosis’; a prominent mode of PCD during host–pathogen interaction.1, 2, 3 While there is still a degree of dispute over the apoptotic roles of inflammatory caspases, the members comprising the other two groups have well-documented functions in the mediation of the apoptotic response.1, 2, 3, 4, 5 Although caspase functions have been a commonly pursued research subject for decades, new properties are continuously attributed to these molecules.3, 6 For instance, caspases owe half their name to their highly specific cleavage of their substrates on aspartate-containing recognition sites. Nevertheless, they have been recently reported to cleave their targets almost as efficiently on motifs bearing glutamate or phosphoserine in place of aspartate.6 Despite their initial discovery as cellular assassins, the exponential increase of evidence that followed in support of their additional non-apoptotic functions has served to view caspases under a different scope; that of agents with many faces, contributing to the regulation of cellular processes that often function complimentary, or even oppositely to apoptosis.3, 7, 8 One such example can be gleaned from their involvement in autophagy, a cellular process intricately connected with apoptosis; so much that often perturbations in one pathway considerably impact the other.9, 10, 11, 12, 13
Autophagy (from the Greek for ‘self-eating’), was coined in 1963 by Nobel laureate C. de Duve.14 30 years later, the seminal work conducted by Y. Ohsumi in describing the core machinery of the process in yeast would in turn lead to his award with the 2016 Nobel Prize.15, 16 Autophagy is subdivided into three types (macroautophagy, microautophagy, and chaperone-mediated autophagy),17, 18, 19 of which macroautophagy (hereafter ‘autophagy’ in this text) is the most well-studied. The process is responsible for the turnover of intracellular constituents, in which a portion of the cytosol is sequestered by isolation membranes. This leads to the formation of a double-membrane-layered organelle called the autophagosome, which subsequently fuses with a lysosome to create an autolysosome, a structure with a single limiting membrane wherein its contents are degraded. The residual materials are then recycled back to the cytosol for effective use by the cell.20 In this manner an assortment of intracellular residents including protein aggregates,21, 22 damaged organelles,23, 24 and invading pathogens,25, 26, 27 as well as caspases28, 29 can be effectively cleared from the cytosolic environment. Therefore, autophagy is primarily regarded as a pro-survival mechanism and is triggered in response to a wide array of life-threating stimuli. The nature of its function places the pathway at the kernel of many physiological settings where apoptosis is also involved.30 Their complex interplay is particularly highlighted during physiological processes such as developmental remodeling and cell differentiation,31 as well as aging.32 Moreover, perturbations which impair the efficient removal of protein aggregates and cytotoxic oligomers by autophagy have been reported in a range of neurodegenerative diseases including Alzheimer’s33 and Parkinson’s.34 In addition, it is posited that some cancer cells can utilize autophagy, as a means to prolong their survival.35, 36
Similarly to apoptosis, progression of autophagy follows a tightly supervised outline of events. Many steps in this cascade are controlled by the interactions of several ATGs with non-autophagy exclusive regulators including caspases.3, 9, 10, 13, 37 To gain a clearer overview of the ways caspases can associate with ATGs and modulate autophagy, we first describe the stepwise assembly of the autophagic machinery in mammalian systems.
Overview of Autophagy
Initiation
The ULK-complex is regarded as the apical initiator of autophagy in mammals.38, 39 Its key kinase ULK is the functional homolog of ATG1, with at least four known members in the family (ULK1-4).40 ULK1 and ULK2 are the most frequently studied in autophagy investigations and they are found to form distinct ULK-complexes within cells,39 mainly due to their different binding affinity for another prominent member of the ULK-complex; ATG13.38, 39, 40 Because of their characterized role in autophagy it is these two members that are being implied hereafter when mentioning either ‘ULK’ or ‘ULK-complex’, unless explicitly specified. Altogether with ULK and ATG13, the core of the ULK-complex is further complemented by ATG10141 and the scaffold protein FIP200.42, 43 FIP200 is the functional homolog of yeast ATG17 and binds ULK, indirectly promoting its kinase activity.38 ATG13 facilitates the interaction between ULK and FIP200 and preferentially binds ULK1 over ULK2.38, 39, 40
Activity of the ULK-complex is in turn modulated further upstream by two Ser/Thr kinases, mTOR 1 and 2.44, 45 These kinases associate with several different binding partners in distinct mTOR complexes (mTORC) with respectively distinct properties and signaling pathways.44, 45 An example of the differential response of each complex related also to autophagy, is that activity of mTORC2 remains unaffected by treatment of cells with the autophagy inducer rapamycin, in contrast to mTORC1.46 This is partly attributed to mTORC2 interacting with Rictor in place of Raptor, which associates with mTORC1;46 the binding partner of mTORC1.44, 45, 46 While they both regulate autophagy, the mTORC2-signaling cascade is still poorly defined therefore, the more well-characterized mTORC1-dependent pathway is often utilized to induce autophagy instead.44, 45, 46
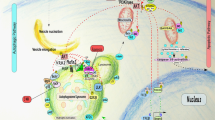
Under fed conditions, mTORC1 binds to and inhibits ULK.47 In contrast, depletion of intracellular amino-acid levels following starvation results in the upregulation of energy level-dependent sensors such as AMPK, which phosphorylates and aids in the dissociation of mTORC from ULK.47, 48 In addition to other post-translational modifications, ULK further activates ATG13, FIP200 and ATG101 within the complex.38, 49 The fully-engaged ULK-complex then translocates to the membrane compartment of phagophore assembly together with the class-III PI3K complex (PI3KC-III); the leading multi-protein structure of the next stage38, 49 (Figure 1).
Overview of the autophagy mechanism in mammalian cells. The proteins are grouped according to their position in the cascade. ‘Core’ contains within its boundaries the key proteins that make up each complex. A. Initiation: On nutrient withdrawal AMPK phosphorylates mTORC1, thereby lifting its inhibitory effect on ULK, and allowing the initiation complex to be activated. ULK phosphorylates AMBRA-1, which subsequently severs the binding between Beclin-1 and Bcl-2 proteins, and facilitates Beclin-1 interaction with p150 and VPS34 in the PI3K-III-complex. UVRAG and ATG14L differentially bind to form distinct PI3K-III-complex populations within cells, while AMBRA-1 also interacts with the PI3K-III-complex. B. Nucleation: The ULK- and PI3K-III- complexes then translocate to the nascent phagophore site with the aid of ATG9 and VMP1, where the localized generation of PI3P leads to the recruitment of additional effectors, and the formation of the initial isolation membrane (not shown). C. Elongation: Shortly after autophagy upregulation, ATG7 and ATG10 are activated and instigate the formation of the ATG5-ATG12 dimer which then binds ATG16L1 to create the ATG16L1 elongation complex. ATG7 also oversees together with ATG4B and ATG3 the cleavage and lipidation of LC3 to LC3-I and then LC3-II, which is bound to the inner and outer membrane of the forming phagophore. LC3-II acts as a recognition marker by LC3-II sensors, which bring cargo targeted for degradation to be sequestered to the autophagosome during its formation, as well as by ATG9 (shown) and other transporter proteins (not shown) which shuttle between various intracellular membrane compartments (for example, mitochondria, Golgi, plasma membrane, shown above) and transfer phospholipid fragments to be incorporated to the expanding structure. D. Maturation: On closure of the loaded phagophore, and the removal of the outer fraction of LC3-II, the now-complete autophagosome can then fuse with a lysosome, through a docking process involving Syntaxin members and the lysosomal LAMP1 and 2 proteins, among others, to create an autolysosome, wherein its contents are degraded by lysosomal hydrolases, and recycled back to the intracellular space to be taken up by numerous effectors (not shown)
Phagophore nucleation and expansion
The lipid kinase VPS34, its regulatory subunit p150, and Beclin-1 (the orthologue of yeast ATG6) are the main components of PI3KC-III, which is essential through the steps of phagophore formation to its maturation to a complete autophagosome.43, 50 Other important co-regulators of the complex include ATG14L, AMBRA-1 and UVRAG.50 ATG14L and UVRAG compete for their binding of Beclin-1 and form separate PI3K-III complexes, which are respectively required in autophagosome biogenesis and in their maturation and fusion with lysosomes.20, 50, 51 Beclin-1 is of particular interest as it sits at the crossroads between autophagy and apoptosis. As a Bcl-2-homology (BH)-3 domain only protein it interacts with BH3-domains of other Bcl-2 family proteins and is often bound inert to the anti-apoptotic Bcl-2 or Bcl-XL proteins at the outer mitochondrial membrane.50 Upon initialization of autophagy, Beclin-1 undergoes phosphorylation and ubiquitination as well as associating with the ULK1-activated scaffold AMBRA-1.50, 52 These changes favor the uncoupling of Beclin-1 from Bcl-2; the freed protein in turn triggers the formation of the PI3K-III-complex by binding and switching-on VPS34.50, 52 At the phagophore nucleation site the upregulated PI3KC-III generates a local pool of PI3Ps, which serve as beacons attracting additional PI3P-sensing phospholipid effectors and ATG proteins on the scene.50 Elongation of the nascent organelle depends on the constant incorporation of membrane material to its expanding body.43 Among the many crucial modulators of this elaborate step, two transmembrane proteins, ATG9 and VMP1 often stand out because of their respective roles. VMP1 tethers PI3KC-III to the developing surface of the phagophore by binding to Beclin-1 in the complex. ATG9 is the only ATG member characterized so far, capable of shuttling between cytosolic compartments and transferring membrane cargo to the developing autophagosome.20
The mammalian Atg8 family consists of 3 subfamilies: LC3, GABARAP and GATE-16.53, 54 Of these, the variants of the LC3-subfamily (LC3A, LC3B, LC3C) and namely LC3B (hereafter LC3), are usual markers to monitor autophagosome formation.53,54 LC3 and the E3-ligase ATG12 are two ubiquitin-like proteins necessary for autophagosome expansion and closure and are recruited to the membrane site along with their associated ubiquitin-like conjugation systems.55, 56 ATG7 activates ATG12 and ATG10 conjugates it to ATG5.9, 50, 55 The resulting ATG12-ATG5 heterodimer then binds ATG16L1, altogether completing the core of the ATG16L1 complex.9, 50, 55 LC3 is cleaved by the cysteine protease ATG4B yielding LC3-I, which is subsequently transferred by ATG7 to the inner and outer membrane of the expanding phagophore.57, 58, 59 There, LC3-I is passed over to ATG3 and by the localized operations of the PI3K-III and ATG16L1 complexes it is lipidated to LC3-II and anchored to the autophagosomal membrane.57, 58, 59
Maturation and fusion with lysosomes
During fusion of the mature autophagosome with a lysosome, the ATG16L1-complex dissociates from the vesicle and ATG4B removes the cytosolic fraction of LC3-II.43, 57, 59
A t-SNARE complex involving several syntaxins and ATG14L generally prime the autophagosome for the fusion step with the lysosome.51 Other reports suggest that an intermediate autophagosome-endosome fusion step might occur, thereby providing the factors necessary for the ensuing lysosomal fusion.60 Acidification and further maturation to fully fledged autolysosomes is mediated with the aid of small GTPases of the Rab family, together with the lysosome-associated-membrane glycoproteins LAMP1 and LAMP2.55, 61
Table 1 summarizes the core ATG and co-regulator proteins discussed here, along with their position in the autophagic cascade and their function.
Caspases Impact on Autophagy
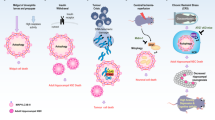
Autophagy and apoptosis are interlocked in an extensive crosstalk with each other, therefore it may not be surprising that many ATGs are recognized and cleaved by caspases.10, 37, 62, 63 In most cases, interaction of the caspase with the ATG protein leads to abrogation of the autophagic function of the latter, and the homeostatic balance shifts in favor of an apoptotic profile (Figure 2).11, 52, 64 This is generally regarded as a commitment of dying cells to an apoptotic PCD by progressively shutting down other processes, including autophagy. Nevertheless, caspase-mediated cleavage of ATGs does not always result in idle waste products; instead several ATG-fragments have been shown to acquire new properties, which can differ greatly from those of their initial full-size isoforms and consequently affect apoptosis and autophagy in numerous ways.65, 66 On the other hand caspases have also been found to promote autophagy under certain contexts.66, 67, 68
Schematic depiction of reported effects of mammalian caspases in autophagy-related effectors. Central red arrow depicts the activation of caspases during the apoptotic cascade for the extrinsic and intrinsic pathway. Following activation of cell-surface expressed death receptors and DISC formation, pro-caspase 8 is converted to caspase-8 in ways which can also involve the ATG16L1-complex, and targeting of pro-caspase-8 oligomers to the autophagosomal surface by p62. Sequestration of pro-caspase-8 to the autophagosome can lead to caspase-8 activation, or downregulation, due to the extensive retention of the protease by the autophagosome, and its consequent degradation by lysosomal hydrolases upon autolysosome formation (not shown). It remains unclear whether pro-caspase-10 is regulated in a similar manner. Initiator caspase-9 is activated during the intrinsic pathway by cytochrome-c release from damaged mitochondria and the assembly of the apoptosome. The initiator caspases then cleave the pro-forms of their effectors, resulting in the upregulation of the executioner caspases -3, -6, and -7, altogether leading to the apoptotic dismantling cells. The side arrows on each caspase depict its known interactions with ATG targets and other autophagy-related proteins. The resulting outcome on autophagy is shown with an ‘X’ over, or a ‘+’ behind the arrows that indicate the otherwise physiological effect of the autophagic protein on the pathway. Alternatively the arrows may point towards other effectors and functions which directly impact autophagy. As shown above the same protein can be targeted by multiple caspases. (Not shown: Caspase-1 interactions, as it is a member of the inflammatory caspase group and associated mostly with the pyroptotic rather than the apoptotic response)
The findings from the existing literature presented below are split between the initiator and the effector caspase groups, pertaining to their main role in apoptosis (as shown in Table 2).
Initiator caspases
Caspase-1 is involved in the maturation of the pro-inflammatory cytokines interleukin-1B/interleukin-18 during oxidative stress.67, 69 In addition, a role of caspase-1 in facilitation of cytoprotective autophagy has been suggested during hypoxia-induced mitochondrial stress.67 In hepatocytes of casp-1 -/- mice, levels of the common autophagy markers LC3 and Beclin-1 were found to be decreased compared to wild-type (WT).67 When Beclin-1 was overexpressed in these cells it was reported that impaired autophagic clearance of damaged mitochondria was restored to WT rates.67 It was therefore suggested that caspase-1 in murine hepatocytes appears to favor autophagy and clearance of damaged mitochondria after redox stress by activating LC3 and Beclin-1 through an as-of-yet undefined mechanism.67
Caspase-2 possess features of both initiator and effector caspases.70 It conveys the apoptotic signal after stimuli such as oxidative stress,71 DNA damage,70 heat shock,72 and cytoskeleton disruption.73 In regards to its involvement in autophagy, under normal conditions caspase-2 is a negative regulator of the process, as supported by results derived from mouse embryonic fibroblasts (MEFs)74 and young adult mouse cortical neurons.71 In both models, casp2 knock-down at an early-stage lead to inhibition of apoptosis and mobilization of cytoprotective autophagy, particularly when the initial death-inducing stimulus was delivered either via heat shock or mitochondrial oxidative stress.71, 74
While the exact mechanism of caspase-2-dependent modulation of autophagy remains to be fully delineated, it seemingly involves control of intracellular levels of reactive oxygen species by caspase-2.71, 74 Loss of caspase-2 favors the accumulation of oxygen radicals, which spark a cascade of events culminating in the downregulation of autophagy suppressors such as TORC and activation of pro-autophagy mediators including AMPK.74
Caspase-8 is involved in the extrinsic apoptotic cascade, shortly after activation of death receptors expressed at the cell-surface.2, 75 DISC is the multi-protein signaling platform assembled on the scene, whereupon the inactive precursor pro-caspase-8 dimerizes and cleaves itself and subsequently released in its functional state to the cytoplasm, triggering upregulation of its effector caspases.75, 76, 77
In addition to its conventional activation pathway, caspase-8 maturation has been reported even in the absence of DISC.78, 79 When formation of this complex is inhibited, autophagy takes over, either as a death-escaping mechanism or as an alternative route for caspase-8 activation.78, 79 Pro-caspase-8 can be localized to autophagosomes via the ATG8/LC3-interacting molecule p62.80 This is an adapter protein which binds poly-ubiquitinated (polyU) protein aggregates and targets them for sequestration at the site of autophagosome formation.81 In this case, the autophagosomal surface may provide a platform where polyU pro-caspase-8 oligomers can be brought in close proximity with each other, thereby facilitating their interaction and maturation to caspase-8 dimers.80, 81, 82, 83 Conversely, it has also been proposed that sequestration of caspase-8 or its precursor to the autophagosome may be a method utilized at least by Bax−/− Hct116 colon carcinoma cells to promote their survival instead.28 It is important to note that these cells develop a resistance to apoptotic stimuli, despite having a fully functional DISC capable of switching on caspase-8.28 However, active caspase-8 and its precursor can still be targeted and retained at the autophagosome where they are eventually degraded along with the rest of the autophagosomal cargo within the autolysosome.28 As there is not enough caspase-8 to adequately propagate the cell death signal, cancer cells become increasingly unresponsive to apoptotic stimuli.28 To rescue caspase-8 from autophagic degradation and re-sensitize cancer cells to apoptotic cell death, a viable option in this instance is to inhibit autophagosome formation.28 As the above examples illustrate, the contradictory functions of the autophagosomal surface in regards to caspase-8 activity should be taken into account in investigative approaches to treat malignant tumors by manipulating autophagy. As this effect seems to be context-dependent it is paramount to find the right balance between promoting targeting of the protease to the developing phagophore and inhibiting autophagy, in order to re-establish an organized apoptotic dismantling of different type of tumor cells. There are already reports where the ability of p62 to promote caspase-8 activation at the autophagosomal membrane has been successfully exploited to render cancerous cell-lines responsive to further pharmacological treatment.83, 84
Moreover, functional caspase-8 has been shown to prevent autophagy from over activation.85 In proliferating T-cells lacking either the DISC-adapter protein FADD or caspase-8, the autophagic response was initially mounted in order to provide the necessary energy to support their proliferation.85 However, the process quickly became hyperactive and led to cell death with necroptotic features such as the upregulation of the kinase RIPK1.85 RIPK1 as well as pro-caspase-8, associate with the ATG16L1 complex, which apart from its autophagosome-expanding role, has been shown to also serve as a scaffold for DISC assembly.80, 85 In WT T-cells, ATG5 is observed to interact with FADD, which also binds both pro-caspase-8 and RIPK1.85 The association of ATG5 with FADD can thereby promote DISC formation and maturation of pro-caspase-8 to active caspase-8 in a manner additionally involving the autophagosome-forming machinery.85 The caspase can then cleave RIPK1, as part of a negative feedback loop to prevent excessive autophagy.85 In the absence of either FADD or caspase-8, RIPK1 remains on and perpetuates the autophagic response, eventually resulting in necroptotic cell death.85 RIPK1 also augmented autophagy in lung, bladder and prostate cancer cell-lines after stimulation of the external apoptotic pathway by TRAIL.86
The role of caspase-8 in modulating autophagy is further complemented by its interactions with ATG3, ATG5 and Beclin-1, which are all reportedly prone to cleavage by the protease in vitro.87, 88 Following death-receptor activation in several cancer cell lines, the subsequent downregulation of autophagic flux has been partly attributed to caspase-8-dependent cleavage of these autophagic proteins.50, 87, 88 A peculiar yet noteworthy finding is that the processing of Beclin-1 by many initiator and effector caspases often returns a C-terminal fragment which in a starvation-induced setting displays increased pro-apoptotic properties.50, 65 Both the extrinsic and intrinsic apoptotic pathway converge at the mitochondrial pro- and anti-apoptotic Bcl-2 proteins, which control the rate of cytochrome-c release.76 The truncated Beclin-1 fragment has been observed to also localize to mitochondria and further promote MOMP and the release of pro-apoptotic factors to the cytosol, consequently amplifying the apoptotic response.50, 65 The processing of ATG3 by caspase-8 was shown to effectively inhibit assembly of the autophagic machinery at an early stage and promote apoptosis.88
Caspase 9 is set in motion following cytochrome c-mediated assembly of the APAF1-apoptosome during the intrinsic apoptotic cascade.2, 89 In regards to the interaction of caspase-9 with autophagic proteins, in several tumor cells caspase 9 cleaves and inactivates to a certain degree ATG5 and Beclin-1.87, 90 In MCF-7 cells in particular, the non-steroidal anti-inflammatory drug FR122047 (FR) is used to induce cell death via the extrinsic apoptotic signaling.91 When MCF-7 cells deficient for caspase-9 were treated with FR, their susceptibility to cellular demise was increased, with a concomitant decrease in autophagic flux.91 The authors postulated that caspase-9 possibly contributes to upregulation of cytoprotective autophagy by mediating proper acidification of lysosomes and the acid-dependent functions of cathepsins.91 As a result, caspase-9-deficient MCF7-cells appear to respond more positively to treatment with FR, due to their inability to mount a proper autophagic response.91
In addition, it was shown in tumor cell-lines as well as MEFs with inducible knock-outs for an array of ATGs, that caspase-9 forms a stable complex with ATG7.68 This interaction does not result in cleavage of ATG7 but conversely the caspase-9-ATG7 complex is more efficient at binding and priming LC3, resulting in a corresponding increase of developing autophagosomes.68 This is partly due to the fact that catalytic activity of caspase-9 while bound to ATG7 is abolished, leaving ATG7 affinity for LC3 unobstructed, thus leading the authors to speculate that this mechanism may be utilized by tumor cells in order to prolong their survival.68
Caspase 10 is activated in an analogous manner to caspase-8 during the extrinsic apoptotic pathway. In addition to its reported cleavage of ATG5 and Beclin-1,50, 87, 90 caspase- 10 has also been discovered to modulate autophagic flux by processing BCLAF1 in a myeloma cell-line.92 BCLAF1 is a known inducer of autophagy capable of indirectly activating Beclin-1 by antagonistically competing for their binding site at Bcl-2.92 This finding was the result of series of experiments utilizing RNA-interference to knock down an array of target proteins required for cell survival, identifying BCLAF-1 to be a substrate of caspase-10.92 Proteolytic cleavage of BCLAF1 by caspase-10 abolished its function and therefore Beclin-1 remained tethered to Bcl-2.92 By effectively exploiting this mechanism, myeloma cells despite their requirement for a degree of background autophagy, seemingly ensure their survival by keeping the process in check and consequently escape demise.92
Effector caspases
Caspase-3, along with caspases -6 and -7, is the principal effector caspase whose role in autophagy regulation has been investigated the most. The first study to suggest a role of mammalian caspase-3 in autophagy utilized TRAIL-induced apoptosis in HeLa cells, during which Beclin-1 was cleaved in a caspase-dependent manner.90 The research group disclosed that proteolytic processing of Beclin-1 by caspase-3 was found to be sufficient in significantly impairing autophagic flux when an apoptotic-inducing stimulus was applied.90 In contrast either overexpression of Beclin-1 in this cell-line or introduction of a non-cleavable mutant effectively inhibited TRAIL-induced cell death in this cell line.90
Moreover, in murine bone marrow cells that underwent prolonged starvation, the initial autophagic response gradually shifted towards apoptosis.65 During this transition stage Beclin-1 was shown to be cleaved by caspase-3, generating a C-terminal fragment that enhanced cell-death by the mechanism already mentioned.65 ATG5 has also been proposed to be prone to cleavage by caspase-3, consequently contributing to autophagy downregulation.87
Other studies argue that caspase-3 in nutrient-deprived human endothelial cells is able to aid towards the export of maturing LC3-containing autophagic vesicles through the plasma membrane, promoting cell-volume shrinkage, which is an indicative apoptotic marker,93, 94
An isoform of the ATG4 protease, ATG4D, is another substrate for caspase-3 processing.95 Cleavage of ATG4D by caspase-3 has been shown to occur at the N-terminal DEVD63K motif, which is a common caspase-cleavage site.66 This action returns a truncated product (ΔN63K-ATG4D) with enhanced lipidation activity on the Atg8-related family member, GABARAP-L1.66, 95 When caspase-3 cleaved the endogenous ATG4D, its resulting truncated isoform was observed to promote autophagy.95 Conversely, when ΔN63K-ATG4D was overexpressed in these cells it was shown to contribute to apoptosis instead.95 This cytotoxic effect is possibly due to the combination of increased levels of ΔN63K-ATG4D and its enhanced affinity for damaged mitochondria where it augments with a currently undefined mechanism, MOMP and the subsequent release of pro-apoptotic factors in the cytosol.66
Furthermore, AMBRA-1, which favors Beclin-1 binding to PI3KC-III, has being identified as an additional target for caspase-3-processing during apoptosis in vitro.52 When AMBRA-1 was inactivated due to proteolytic cleavage by caspase-3, Beclin-1 consequently failed to couple effectively to PI3KC-III and autophagosome formation was inhibited as a net result.52 Autophagy was thereby halted in an AMBRA-1-dependent fashion, favoring the progress of the apoptotic cascade.52
Caspase 6 impact on autophagy progression has been suggested, along with other caspases, based on observations from cancer-derived cell cultures.87, 90 In melanoma-derived cell-lines subjected to starvation and TRAIL-induced cell death, caspase-6 was shown to exacerbate degradation of Beclin-1 and ATG5, further inhibiting autophagy and shifting the balance in favor of an apoptotic response in these cells.87
Moreover, an in vitro screening for identifying protease regulators of ATG proteins in mammalian cells showed that also ATG3 and p62 are potential substrates for caspase-6 processing.62 When ATG3 is cleaved by the effector caspase, autophagy is subsequently downregulated, possibly due to inhibition of phagophore formation.62 However, the exact reason why the pathway is inhibited as a result of p62 loss is yet unclear. A possible explanation might lie in the role of p62 as a promoter of autophagy by targeting LC3 and polyU-aggregates at the site of autophagosome formation.82, 96 Its loss due to caspase-6 cleavage may therefore result in the less efficient recruitment of LC3 and polyU-cargo to the autophagosome, inevitably posing a setback in autophagy progression.62 In a potential extension of this argument, in several settings p62 can modulate caspase-8 function by targeting its polyU-precursor to the autophagosome during apoptosis upregulation, as already mentioned.82, 83 Caspase-8 then activates downstream executioners such as caspase-6, thereby enhancing the apoptotic response.1 As such, it may be posited that in those same settings caspase-6 may potentially cleave p62 as part of a negative feedback loop that further prohibits the recruitment of LC3 and polyU-cargo to autophagosomes.
Caspase-7 is the lesser known in regards to its impact on the autophagic pathway. A study demonstrated the ability of caspase-7 to bind and cleave a mutated form of ATG16L1 (mutATG16L1).97 Binding affinity of caspase-7 was greater for mutATG16L1 compared to WT ATG16L1.97 In humans, this single coding polymorphism is associated with increased risk for developing Crohn’s disease,97 and caspase-7 binding to mutATG16L1 was correlated to an observed decrease of autophagy in response to pathogen-infection.97 The group concluded that even though the exact mechanism warrants further investigation, compromised cytoprotective autophagy in this context appears to be associated with increased bacterial load, further disrupting cellular integrity and potentially contributing to the disease manifesting in cells.97
Caspase-Autophagy Cross-Regulation in Developmental Cell Death in Drosophila
The role of autophagy as a cell death modality has sparked increased controversy among researchers. However, it is agreed that since it is often difficult to distinguish whether autophagy is a causal or an auxiliary phenomenon of cell death, the line between ‘cell death by autophagy’ and ‘cell death with autophagy’ must be drawn as clearly as possible in functional studies,98, 99 There are excellent reviews available that deal in greater depth with this condition-specific duality of the pathway.12, 100
The fruit fly Drosophila has provided substantial evidence regarding the role of autophagy as a cell-death mediator during developmental remodeling.101, 102 Upon salivary gland removal in late larval stages, autophagy functions in parallel with caspase-dependent apoptosis to degrade the larval tissue.103 Mutations of single key Atg genes or inhibition of the Drosophila initiator caspase DRONC, severely delay this degradation step.103 Restoring expression of Atg1 in these cells re-vitalizes the autophagic response and leads to premature cell death in a caspase-independent fashion.103
Furthermore, upon larvae to pupae transition stage, midgut cells of the intestine are disposed in a manner predominantly driven by autophagy and secondarily by apoptosis.104, 105 In this setting, inhibition of core Atgs has been observed to negatively influence the rate of midgut cell degradation, which in contrast to salivary glands is not delayed further by inhibiting individual caspases or by using the pan-caspase inhibitor p35.103, 105
During Drosophila oogenesis, degradation of supporting nurse cells by PCD, has been shown to promote the maturation of the oocyte.106, 107, 108 In nurse cells the effector caspase DCP-1 (the Drosophila homolog to caspase-3) and its inhibitor of apoptosis dBRUCE were firstly observed to regulate autophagic flux, which contributed in turn to ovarian PCD.109 Shortly after, it was shown that activation of PCD relied on the apical degradation of dBRUCE by autophagy, which enhanced DCP-1 function and autophagy upregulation in a positive feedback loop, involving DCP-1 translocation to mitochondria and its localized suppression of a recently identified autophagy-inhibitor.107, 110 Nevertheless, since the effective removal of nurse cells has also been reported even when inhibiting both apoptosis and autophagy,111 the conditions under which autophagy functions as or contributes to death-mediating pathways require further elucidation.
Conclusions
The precise mechanisms by which caspases manage to regulate autophagy remain largely unknown. It is important to note that unless the initial stress-inducing stimulus is overwhelming, most cells will firstly aim to survive by mounting an autophagic response, before resorting to suicide.9, 10, 13 In spite of its cytoprotective role, if autophagy progresses unimpeded over and above a certain threshold, it can lead to upregulation of cell-death markers and activation of the apoptotic machinery or other cell death modalities if apoptosis is impaired.13 In this regard, some caspases can evidently act as molecular brakes by degrading ATGs as a means to keep the process from reaching a point of no return, past which the cellular suicide machineries are mobilized. This is particularly exemplified with apical caspase-8, as it switches off the autophagy-enhancer RIPK1, therefore keeping the autophagic flux in check.85 In contrast to this, the non-apoptotic role of caspase-9 appears to be mostly a matter of its limited availability due to its association with ATG7 in a complex, which nonetheless favors autophagy upregulation.68 Furthermore, the differential proteolytic cleavage of autophagy-proteins by initiator and executioner caspases can result in fragments, which display either pro-apoptotic or pro-autophagic properties as discussed for Beclin-1 and ATG4D respectively.
The gradual unveiling of hidden functions for known regulatory proteins also births new questions. Is caspase-control of autophagy an essential mode for managing this pathway? Caspase involvement stands out particularly in apoptosis-inducing settings. As autophagy is a highly elaborate pathway, many signaling networks converge to regulate the process, therefore it is reasonable to expect that depending on setting, certain networks may impact the pathway more prominently than others. Do the caspase precursors have any type of control over the autophagic cascade? Perhaps. As the modulatory effects of pro-caspases in relation to autophagy is a largely unexplored avenue, it remains to be seen if and to what extent they can potentially affect the process. On the other hand, as already reported for pro-caspase-8, the autophagic machinery can be indispensable as an alternative path to the maturation of the protease in conditions where the conventional mode of activation is blocked.78, 79 Whether other cell-death proteases can be upregulated in a similar manner is only one of the many questions to be answered in the future. Since caspases have been known to contribute to processes such as cell-proliferation and differentiation, which are contradictory to apoptosis,3, 112, 113 it is possible that these misunderstood hitmen may have additional roles in autophagy awaiting discovery. Taking all the presented findings into account, the primary conclusions of the review are that caspases can directly interact with known ATGs and consequently affect autophagy in a number of ways. In the majority of cases autophagy is downregulated in favor of apoptosis. Interestingly enough, apical caspases -1, -8, and -9, as well as executioner caspase-3 have been found to promote autophagy on occasions. Overall, the net effect on autophagy due to caspase-ATG interactions appears to rely heavily on context, and further work is required to delineate the complex relationships between autophagic proteins and cell death proteases in health and disease.
References
McIlwain DR, Berger T, Mak TW . Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol 2013; 5: a008656–a008656.
Parrish AB, Freel CD, Kornbluth S . Cellular mechanisms controlling caspase activation and function. Cold Spring Harb Perspect Biol 2013; 5: a008672.
Shalini S, Dorstyn L, Dawar S, Kumar S . Old, new and emerging functions of caspases. Cell Death Differ 2014; 22: 526–539.
Abraha AM, Ketema EB . Apoptotic pathways as a therapeutic target for colorectal cancer treatment. World J Gastrointest Oncol 2016; 8: 583–591.
Wong RSY . Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res 2011; 30: 87.
Seaman JA, Julien JE, Lee O, Rettenmaeier PS, Thomsen TJ, Wells ND . Cacidases: caspases can cleave after aspartate, glutamate, and phosphoserine residues. Cell Death Differ 2016; 23: 1717–1726.
Hyman BT, Yuan J . Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nat Rev Neurosci 2012; 13: 395–406.
Kuranaga E, Miura M . Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol 2007; 17: 135–144.
Rubinstein AD, Kimchi A . Life in the balance – a mechanistic view of the crosstalk between autophagy and apoptosis. J Cell Sci 2012; 125: 5259–5268.
Booth LA, Tavallai S, Hamed HA, Cruickshanks N, Dent P . The role of cell signalling in the crosstalk between autophagy and apoptosis. Cell Signal 2014; 26: 549–555.
Oral O, Akkoc Y, Bayraktar O, Gozuacik D . Physiological and pathological significance of the molecular cross-talk between autophagy and apoptosis. Histol Histopathol 2016; 31: 479–498.
Anding AL, Baehrecke EH . Autophagy in cell life and cell death. Curr Top Dev Biol 2015; 114: 67–91.
Marino G, Niso-Santano M, Baehrecke EH, Kroemer G . Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014; 15: 81–94.
Klionsky DJ . Autophagy revisited: a conversation with Christian de Duve. Autophagy 2008; 4: 740–743.
Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y . Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 1992; 119: 301–311.
Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD et al. A protein conjugation system essential for autophagy. Nature 1998; 395: 395–398.
Li WW, Li J, Bao JK . Microautophagy: lesser-known self-eating. Cell Mol Life Sci 2012; 69: 1125–1136.
Kaushik S, Cuervo AM . Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol 2012; 22: 407–417.
Parzych KR, Klionsky DJ . An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 2014; 20: 460–473.
He C, Klionsky DJ . Regulation Mechanisms and Signalling Pathways of Autophagy. Annu Rev Genet 2009; 43: 67.
Lamark T, Johansen T . Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int J Cell Biol 2012; 2012: 1–21.
Wong E, Bejarano E, Rakshit M, Lee K, Hanson HH, Zaarur N et al. Molecular determinants of selective clearance of protein inclusions by autophagy. Nat Commun 2012; 3: 1240.
Zhang J . Autophagy and mitophagy in cellular damage control. Redox Biol 2013; 1: 19–23.
Narendra D, Tanaka A, Suen DF, Youle RJ . Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008; 183: 795–803.
Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A . Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007; 315: 1398–1401.
Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH . Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem 2006; 281: 11374–11383.
Deretic V, Saitoh T, Akira S . Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013; 13: 722–737.
Hou W, Han J, Lu C, Goldstein LA, Rabinowich H . Autophagic degradation of active caspase-8: A crosstalk mechanism between autophagy and apoptosis. Autophagy 2010; 6: 891–900.
Zhang YB, Zhao W, Zeng RX . Autophagic degradation of Caspase-8 protects U87MG cells against H2O2-induced oxidative stress. Asian Pacific J Cancer Prev 2013; 14: 4095–4099.
Mizushima N . Physiological functions of autophagy. Curr Top Microbiol Immunol 2009; 335: 71–84.
Mizushima N, Levine B . Autophagy in mammalian development and differentiation. Nat Cell Biol 2010; 12: 823–830.
Rubinsztein DC, Mariño G, Kroemer G . Autophagy and aging. Cell 2011; 146: 682–695.
Nilsson P, Loganathan K, Sekiguchi M, Matsuba Y, Hui K, Tsubuki S et al. Aβ secretion and plaque formation depend on autophagy. Cell Rep 2013; 5: 61–69.
Lynch-Day MA, Mao K, Wang K, Zhao M, Klionsky DJ . The role of autophagy in Parkinson’s disease. Cold Spring Harb Perspect Med 2012; 2: a009357.
Mathew R, Karantza-Wadsworth V, White E . Role of autophagy in cancer. Nat Rev Cancer 2007; 7: 961–967.
Kenific CM, Debnath J . Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol 2015; 25: 37–45.
Wu H, Che X, Zheng Q, Wu A, Pan K, Shao A et al. Caspases: a molecular switch node in the crosstalk between autophagy and apoptosis. Int J Biol Sci 2014; 10: 1072–1083.
Jung CH, Jun CB, Ro S-H, Kim Y-M, Otto NM, Cao J et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009; 20: 1992–2003.
Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X . ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009; 284: 12297–12305.
Jung CH, Ro SH, Cao J, Otto NM, Kim DH . MTOR regulation of autophagy. FEBS Lett 2010; 584: 1287–1295.
Mercer CA, Kaliappan A, Dennis PB . A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 2009; 5: 649–662.
Hara T, Takamura A, Kishi C, Iemura S-II, Natsume T, Guan J-LL et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008; 181: 497–510.
Lamb CA, Yoshimori T, Tooze SA . The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 2013; 14: 759–774.
Kim LC, Cook RS, Chen J . mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 2017; 36: 2191–2201.
Zoncu R, Efeyan A, Sabatini DM . mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12: 21–35.
Gaubitz C, Prouteau M, Kusmider B, Loewith R . TORC2 structure and function. Trends Biochem Sci 2016; 41: 532–545.
Alers S, Loffler AS, Wesselborg S, Stork B . Role of AMPK-mTOR-Ulk1/2 in the Regulation of Autophagy: Cross Talk, Shortcuts, and Feedbacks. Mol Cell Biol 2012; 32: 2–11.
Ulgherait M, Rana A, Rera M, Graniel J, Walker DW . AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep 2014; 8: 1767–1780.
McEwan DG, Dikic I . The Three Musketeers of Autophagy: Phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 2011; 21: 195–201.
Kang R, Zeh HJ, Lotze MT, Tang D . The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18: 571–580.
Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015; 520: 563–566.
Pagliarini V, Wirawan E, Romagnoli A, Ciccosanti F, Lisi G, Lippens S et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ 2012; 19: 1495–1504.
Shpilka T, Weidberg H, Pietrokovski S, Elazar Z . Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 2011; 12: 226.
Behrends C, Sowa ME, Gygi SP, Harper JW . Network organization of the human autophagy system. Nature 2010; 466: 68–76.
Chua CEL, Gan BQ, Tang BL . Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci 2011; 68: 3349–3358.
Geng J, Klionsky DJ . The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 2008; 9: 859–864.
Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E . HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem 2004; 279: 36268–36276.
Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N et al. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408: 488–492.
Bortnik S, Choutka C, Horlings HM, Leung S, Baker JHE, Lebovitz C et al. Identification of breast cancer cell subtypes sensitive to ATG4B inhibition. Oncotarget 2014; 7: 66970–66988.
Mizushima N . Autophagy: process and function. Genes Dev 2007; 21: 2861–2873.
Huynh KK, Eskelinen E-L, Scott CC, Malevanets A, Saftig P, Grinstein S . LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J 2007; 26: 313–324.
Norman JM, Cohen GM, Bampton ETW . The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy 2010; 6: 1042–1056.
Djavaheri-Mergny M, Maiuri MC, Kroemer G . Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010; 29: 1717–1719.
Zhu Y, Zhao L, Liu L, Gao P, Tian W, Wang X et al. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell 2010; 1: 468–477.
Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 2010; 1: e18.
Betin VMS, Lane JD . Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci 2009; 122: 2554–2566.
Sun Q, Gao W, Loughran P, Shapiro R, Fan J, Billiar TR et al. Caspase 1 activation is protective against hepatocyte cell death by up-regulating beclin 1 protein and mitochondrial autophagy in the setting of redox stress. J Biol Chem 2013; 288: 15947–15958.
Han J, Hou W, Goldstein LA, Stolz DB, Watkins SC, Rabinowich H . A complex between Atg7 and caspase-9: A novel mechanism of cross-regulation between autophagy and apoptosis. J Biol Chem 2014; 289: 6485–6497.
Fink SL, Cookson BT . Apoptosis, pyroptosis, and necrosis: Mechanistic description of dead and dying eukaryotic cells. Infect Immun 2005; 73: 1907–1916.
Zhivotovsky B, Orrenius S . Caspase-2 function in response to DNA damage. Biochem Biophys Res Commun 2005; 331: 859–867.
Tiwari M, Lopez-Cruzan M, Morgan WW, Herman B . Loss of caspase-2-dependent apoptosis induces autophagy after mitochondrial oxidative stress in primary cultures of young adult cortical neurons. J Biol Chem 2011; 286: 8493–8506.
Tu S, McStay GP, Boucher L-M, Mak T, Beere HM, Green DR . In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nat Cell Biol 2006; 8: 72–77.
Ho LH, Read SH, Dorstyn L, Lambrusco L, Kumar S . Caspase-2 is required for cell death induced by cytoskeletal disruption. Oncogene 2008; 27: 3393–3404.
Tiwari M, Sharma LK, Vanegas D, Callaway DA, Bai Y, Lechleiter JD et al. A nonapoptotic role for CASP2/caspase 2: Modulation of autophagy. Autophagy 2014; 10: 1054–1070.
Zhao Y, Sui X, Hong R . From procaspase-8 to caspase-8: Revisiting structural functions of caspase-8. J Cell Physiol 2010; 225: 316–320.
Gross A, Yin XM, Wang K, Wei MC, Jockel J, Milliman C et al. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-X(L) prevents this release but not tumor necrosis factor-R1/Fas death. J Biol Chem 1999; 274: 1156–1163.
Ferreira KS, Kreutz C, MacNelly S, Neubert K, Haber A, Bogyo M et al. Caspase-3 feeds back on caspase-8, Bid and XIAP in type I Fas signaling in primary mouse hepatocytes. Apoptosis 2012; 17: 503–515.
Zhu K, Dunner K, McConkey DJ . Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene 2010; 29: 451–462.
Laussmann Ma, Passante E, Düssmann H, Rauen Ja, Würstle ML, Delgado ME et al. Proteasome inhibition can induce an autophagy-dependent apical activation of caspase-8. Cell Death Differ 2011; 18: 1584–1597.
Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem 2012; 287: 12455–12468.
Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171: 603–614.
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-AA, Outzen H et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–24145.
Huang S, Okamoto K, Yu C, Sinicrope FA . P62/sequestosome-1 Up-regulation promotes ABT-263-induced caspase-8 aggregation/activation on the autophagosome. J Biol Chem 2013; 288: 33654–33666.
Pan JA, Fan Y, Gandhirajan RK, Madesh M, Zong WX . Hyperactivation of the mammalian degenerin MDEG promotes caspase-8 activation and apoptosis. J Biol Chem 2013; 288: 2952–2963.
Bell BD, Leverrier S, Weist BM, Newton RH, Arechiga AF, Luhrs Ka et al. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci USA 2008; 105: 16677–16682.
He W, Wang Q, Xu J, Xu X, Padilla MT, Ren G et al. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2- And RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy 2012; 8: 1811–1821.
You M, Savaraj N, Kuo MT, Wangpaichitr M, Varona-Santos J, Wu C et al. TRAIL induces autophagic protein cleavage through caspase activation in melanoma cell lines under arginine deprivation. Mol Cell Biochem 2013; 374: 181–190.
Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, Gozuacik D . Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis 2012; 17: 810–820.
Würstle ML, Laussmann MA, Rehm M . The central role of initiator caspase-9 in apoptosis signal transduction and the regulation of its activation and activity on the apoptosome. Exp Cell Res 2012; 318: 1213–1220.
Cho DH, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC . Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett 2009; 274: 95–100.
Jeong HS, Choi HY, Lee ER, Kim JH, Jeon K, Lee HJ et al. Involvement of caspase-9 in autophagy-mediated cell survival pathway. Biochim Biophys Acta - Mol Cell Res 2011; 1813: 80–90.
Lamy L, Ngo VN, Emre NCT, Shaffer AL, Yang Y, Tian E et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell 2013; 23: 435–449.
Sirois I, Groleau J, Pallet N, Brassard N, Hamelin K, Londono I et al. Caspase activation regulates the extracellular export of autophagic vacuoles. Autophagy 2012; 8: 927–937.
Nunez R, Sancho-Martinez SM, Novoa JML, Lopez-Hernandez FJ . Apoptotic volume decrease as a geometric determinant for cell dismantling into apoptotic bodies. Cell Death Differ 2010; 17: 1665–1671.
Betin VMS, Lane JD . Atg4D at the interface between autophagy and apoptosis. Autophagy 2009; 5: 1057–1059.
Johansen T, Lamark T . Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011; 7: 279–296.
Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci USA 2014; 111: 7741–7746.
Denton D, Nicolson S, Kumar S . Cell death by autophagy: facts and apparent artefacts. Cell Death Differ 2012; 19: 87–95.
Kroemer G, Levine B . Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol 2008; 9: 1004–1010.
Denton D, Xu T, Kumar S . Autophagy as a pro-death pathway. Immunol. Cell Biol. 2014; 93: 35–42.
Zirin J, Perrimon N . Drosophila as a model system to study autophagy. Semin Immunopathol 2010; 32: 1–10.
Mulakkal NC, Nagy P, Takats S, Tusco R, Juhász G, Nezis IP . Autophagy in drosophila: From historical studies to current knowledge. Biomed Res Int 2014; 2014: 273473.
Berry DL, Baehrecke EH . Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 2007; 131: 1137–1148.
Denton D, Shravage B, Simin R, Baehrecke EH, Kumar S . Larval midgut destruction in Drosophila: Not dependent on caspases but suppressed by the loss of autophagy. Autophagy 2010; 6: 163–165.
Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH et al. Autophagy, Not Apoptosis, Is Essential for Midgut Cell Death in Drosophila. Curr Biol 2009; 19: 1741–1746.
Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjørkøy G, Johansen T et al. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy 2009; 5: 298–302.
Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjørkøy G, Johansen T et al. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol 2010; 190: 523–531.
Peterson JS, Timmons AK, Mondragon AA, McCall K . The end of the beginning: Cell death in the Germline. Curr Top Dev Biol 2015; 114: 93–119.
Hou YCC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM . Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol 2008; 182: 1127–1139.
DeVorkin L, Go NE, Hou YCC, Moradian A, Morin GB, Gorski SM . The Drosophila effector caspase Dcp-1 regulates mitochondrial dynamics and autophagic flux via SesB 2014 J Cell Biol 205: 477–492.
Peterson JS, McCall K . Combined inhibition of autophagy and caspases fails to prevent developmental nurse cell death in the Drosophila melanogaster ovary. PLoS ONE 2013; 8: e76046.
Miura M . Apoptotic and nonapoptotic Caspase function in animal development. Cold Spring Harb Perspect Biol 2016; 4: a008664.
Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G . Non-apoptotic functions of apoptosis-regulatory proteins. EMBO Rep 2012; 13: 322–330.
Acknowledgements
This work is supported by BBSRC grants BB/L006324/1 and BB/P007856/1 awarded to I.P.N.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by E Arama
Rights and permissions
About this article
Cite this article
Tsapras, P., Nezis, I. Caspase involvement in autophagy. Cell Death Differ 24, 1369–1379 (2017). https://doi.org/10.1038/cdd.2017.43
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2017.43
This article is cited by
-
Resveratrol and quercetin protect from Benzo(a)pyrene-induced autophagy in retinal pigment epithelial cells
International Ophthalmology (2024)
-
Cadmium toxicity and autophagy: a review
BioMetals (2024)
-
Apoptosis and eryptosis: similarities and differences
Apoptosis (2024)
-
Moschus ameliorates glutamate-induced cellular damage by regulating autophagy and apoptosis pathway
Scientific Reports (2023)
-
Enhancement of Immunosuppressive Activity of Mesenchymal Stromal Cells by Platelet-Derived Factors is Accompanied by Apoptotic Priming
Stem Cell Reviews and Reports (2023)