Abstract
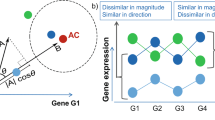
A central goal of gene expression studies coupled with drug response screens is to identify predictive profiles that can be exploited to stratify patients. Numerous methods have been proposed towards this end, most of them focusing on novel statistical methods and model selection techniques that attempt to uncover groups of genes, whose expression profiles are directly and robustly correlated with drug response. However, biological systems process information through the crosstalk of multiple signaling networks, whose ultimate phenotypic consequences may only be determined by the combined input of relevant interacting systems. By restricting predictive signatures to direct gene–drug correlations, biologically meaningful interactions that may serve as superior predictors are ignored. Here we demonstrate that predictive signatures, which incorporate the interaction between background gene expression patterns and individual predictive probes, can provide superior models than those that directly relate gene expression levels to pharmacological response, and thus should be more widely utilized in pharmacogenetic studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Shoemaker RH . The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006; 6: 813–823.
Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci USA 2001; 98: 10787–10792.
Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med 2006; 12: 1294–1300.
Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci USA 2007; 104: 13086–13091.
Butte AJ, Tamayo P, Slonim D, Golub TR, Kohane IS . Discovering functional relationships between RNA expression and chemotherapeutic susceptibility using relevance networks. Proc Natl Acad Sci U S A 2000; 97: 12182–12186.
Nevins JR, Potti A . Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet 2007; 8: 601–609.
Schadt EE, Friend SH, Shaywitz DA . A network view of disease and compound screening. Nat Rev Drug Discov 2009; 8: 286–295.
Fambrough D, McClure K, Kazlauskas A, Lander ES . Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell 1999; 97: 727–741.
Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR . The p53 network. J Biol Chem 1998; 273: 1–4.
Bai X, Jiang Y . Key factors in mTOR regulation. Cell Mol Life Sci 2010; 67: 239–253.
Holter NS, Mitra M, Maritan A, Cieplak M, Banavar JR, Fedoroff NV . Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proc Natl Acad Sci U S A 2000; 97: 8409–8414.
Alter O, Brown PO, Botstein D . Singular value decomposition for genome-wide expression data processing and modeling 2000. Proc Natl Acad Sci U S A 97: 10101–10106.
Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N . Revealing modular organization in the yeast transcriptional network. Nat Genet 2002; 31: 370–377.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D . Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Patterson N, Price AL, Reich D . Population structure and eigenanalysis. PLoS Genet 2006; 2: e190.
Yang JJ, Cheng C, Devidas M, Cao X, Fan Y, Campana D et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet 2011; 43: 237–241.
Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther 2007; 6: 820–832.
Rajaram S, Oono Y . 2010. NeatMap—non-clustering heat map alternatives in R. BMC Bioinformatics 2010; 11: 45.
Engreitz JM, Daigle Jr BJ, Marshall JJ, Altman RB . Independent component analysis: mining microarray data for fundamental human gene expression modules. J Biomed Inform 2010; 43: 932–944.
Kitano H . Systems biology: a brief overview. Science 2002; 295: 1662–1664.
Chow WA, Jiang C, Guan M . Anti-HIV drugs for cancer therapeutics: back to the future? Lancet Oncol 2009; 10: 61–71.
Li Y, Lin AW, Zhang X, Wang Y, Wang X, Goodrich DW . Cancer cells and normal cells differ in their requirements for Thoc1. Cancer Res 2007; 67: 6657–6664.
Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 2009; 137: 835–848.
Cocconi G, Bella M, Calabresi F, Tonato M, Canaletti R, Boni C et al. Treatment of metastatic malignant melanoma with dacarbazine plus tamoxifen. N Engl J Med 1992; 327: 516–523.
Qin Y, Yao L, King EE, Buddavarapu K, Lenci RE, Chocron ES et al. Germline mutations in TMEM127 confer susceptibility to pheochromocytoma. Nat Genet 2010; 42: 229–233.
Boulay A, Rudloff J, Ye J, Zumstein-Mecker S, O’Reilly T, Evans DB et al. Dual inhibition of mTOR and estrogen receptor signaling in vitro induces cell death in models of breast cancer. Clin Cancer Res 2005; 11: 5319–5328.
Acknowledgements
This work was supported by an NIH Center for Translational Science Award UL1 RR025774 (AT, NJS). NJS is supported by the following grants: U19 AG023122-05, R01 MH078151-03, N01 MH22005, U01 DA024417-01, P50 MH081755-01, R01 AG030474-02, N01 MH022005, R01 HL089655-02, R01 MH080134-03, U54 CA143906-01.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Torkamani, A., Schork, N. Background gene expression networks significantly enhance drug response prediction by transcriptional profiling. Pharmacogenomics J 12, 446–452 (2012). https://doi.org/10.1038/tpj.2011.35
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tpj.2011.35
Keywords
This article is cited by
-
Drug response prediction using graph representation learning and Laplacian feature selection
BMC Bioinformatics (2022)
-
A multi-site feasibility study for personalized medicine in canines with Osteosarcoma
Journal of Translational Medicine (2013)